Ants, Bees, Genomes & Evolution @ Queen Mary University London
Published: 09 March 2010
Parasitoid Wasps: From Natural History to Genomic Studies
Yannick Wurm, Laurent Keller
Current Biology, 2010, 5:R242-4
Summary
The sequencing of three Nasonia genomes provides new insights on the molecular signature associated with parasitoid lifestyle, allows comparison with the social honey bee, and enables the identification of genes underlying between-species and sex-specific differences.
Main Text
“I cannot persuade myself that a beneficent and omnipotent God would have designedly created the Ichneumonidae with the express intention of their feeding within the living bodies of caterpillars.”
Charles Darwin, May 22nd, 1860
With these words to his theist friend, the renowned botanist Asa Gray, Darwin expressed his astonishment at the extremely specialized and selfish lifestyles exhibited by parasitoid wasps. For example, adult females of the jewel wasp Nasonia vitripennis locate pupae of filth fly hosts, drill through the pupae’s exoskeleton, inject a potent venom and deposit a few dozen eggs. The young feed on the paralyzed host until development is complete. After eclosion, males do not disperse and typically mate with their sisters. This unusual mating system has attracted much attention by naturalists and evolutionary biologists, in particular because it allows quantitative tests of sex ratio evolution and adaptation [1, 2, 3, 4]. The sequencing of three Nasonia genomes [5]now opens new doors to study many aspects of Nasonia’s life history in molecular and genetic terms, as attested by the publication of more than thirty companion papers.
Several striking features, some probably associated with the parasitoid lifestyle, mark the Nasonia genome. First, the odorant binding protein family, likely to be involved in the selective detection of hosts and the avoidance of toxic elements around filth-dwelling hosts [5, 6], has undergone genetic expansion and shows evidence for rapid turnover. Second, Nasonia exhibits an unequaled richness of venom proteins, providing means to manipulate the host, arrest its development and prevent immune responses [7]. Third, Nasonia carries unusually high amounts of repetitive DNA of all sorts, including transposable elements, nuclear-mitochondrial insertions and microsatellites [5]. This is somewhat surprising given that transposable elements often have deleterious effects and Nasonia males are haploid, which should allow for more effective purging of deleterious mutations. The large amount of repetitive DNA is perhaps due to the genetic drift associated with bottlenecks and inbreeding that are prominent features of Nasonia and many other parasitoids. Consistent with the view that effective population sizes of Nasonia species are relatively small, comparative studies of several strains and populations showed low genetic variability [5, 8].
After the honey bee, Nasonia is the second hymenopteran to have its genome sequenced, thus providing a good opportunity to test whether some of the peculiarities of the honey bee genome are really due to the evolution of social life. The examination of the evolutionary rates of three groups of honey bee genes revealed differences in the rate of evolution of genes depending on caste-specific differences in expression. Queen-specific genes are apparently evolving faster, followed by genes that are neither queen nor worker-specific and finally worker-specific genes evolve at the slowest rates [9]. This result is consistent with the prediction that genes under direct selection (queen-specific and non caste-specific genes) evolve faster than genes under indirect selection (genes expressed only in non-reproductive workers) [10].
Surprisingly, evolutionary rates of the Nasonia orthologs of the three groups of genes show the same pattern [9]. This suggests that genes that had a faster evolutionary rate in the solitary ancestor of honey bees became queen-specific more frequently than slower-evolving genes. Many of these fast-evolving genes have functions linked to metabolism [9]. Their greater likelihood to become queen-specific perhaps reflects the fact that queens have a much higher reproductive potential than workers. It will be interesting to see whether a similar pattern holds true for social wasps and ants for which molecular-genetic tools are becoming available [11, 12, 13].
The Nasonia genome also sheds some light on the evolution of two particular families of proteins that had been emphasized by the honey bee genome project. The honey-bee-specific expansion of the yellow-related major royal jelly gene subfamily has been suggested to be a result of the evolution of sociality because food for larval queen bees is primarily made up of their protein products [14, 15]. The Nasonia genome, however, exhibits a similar, yet independent, expansion of major royal jelly-like genes [5], suggesting that the honey bee expansion is unrelated to social evolution. A similar surprise comes from the discovery that, like honey bees, Nasonia have approximately half as many immunity genes as the fruit fly, mosquitoes and the flour beetle. In the honey bees, it was suggested that this was either because social life provided a sheltered environment (the hive) and group-level means to fight against diseases or because workers feed on pollen and nectar, which are relatively safe food sources [14, 16]. These explanations are implausible for Nasonia, which is solitary and likely to be subject to a large range of pathogens during parasitism. Clearly, data on the diversity of immune genes from more insects with different life-histories will be needed to understand the selective forces responsible for between-species differences.
The sequencing of the *Nasonia vitripennis* genome, together with the possibility to conduct crosses with three closely-related Nasonia species, two of which have draft genome sequences, provides a unique opportunity to examine genome evolution on short timescales. As Nasonia emerges as a genetic model, quantitative trait loci involved in host identification, sex ratio determination, male courtship and diapause induction have already been mapped to small chromosomal regions [5, 17]. Most interestingly, ws1, a locus responsible for a 45% difference in wing size in males but not females (Figure 1), has been mapped down to a 13,500 basepair non-coding region containing the 5′-untranslated rection and cis-regulatory domain of doublesex [18]. Doublesex, a master sex determination gene found from nematodes to mammals, is likely to play a key role in Nasonia sex determination via the expression of sex-specific spliceforms [19]. The allele-specific increase in male wing size is correlated to higher expression of the male spliceform of doublesex in wing tissue [18]. This suggests that ws1 alleles induce wing-specific differential expression of the mediator of sex differentiation doublesex and the modulation of the wing developmental pathway which ultimately leads to differences in wing size.
Figure 1 – Phenotypes of traits expressed in a sex-specific manner in Nasonia vitripennis (A,B,C) and rock-dwelling cichlid fish from Lake Malawi (D,E,F).
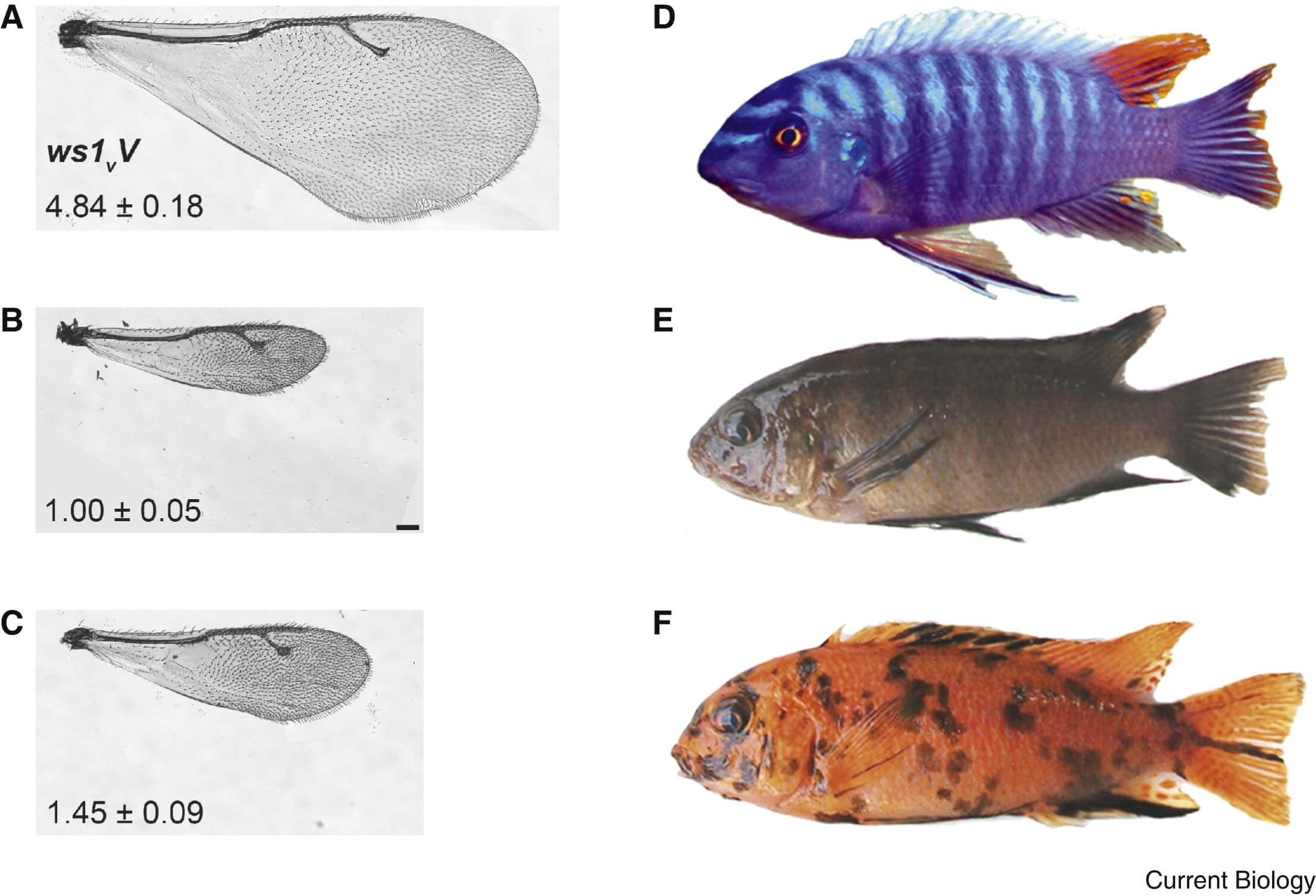
(A) Typical female wing. (B) Typical male N. vitripennis wing. (C) Wing of a N. vitripennis male that carries the 13,500 bp ws1 allele from N. giraulti. (D) Typical male nuptial coloration. (E) Typical non-OB female. (F) Typical OB female.
This type of evolution of sex and tissue-specific expression via regulatory elements linked to a sex differentiation gene may be a common manner of resolving genetic conflicts resulting from sexually antagonistic selection. For example, a recent study [20] of rock-dwelling Lake Malawi cichlid fish (Figure 1) showed that the orange-blotch (OB) locus, which apparently leads to increased female but decreased male fitness, contains Pax7, a gene known to specify the fate of pigmentation cells. The OB allele at Pax7 shows two-fold higher expression than the non-OB allele but both have the same coding sequences, suggesting that cis-regulatory differences account for the OB phenotype. Additionally, all fish carrying the OB allele are female while non-OB fish can be of either sex. This indicates that OB is linked to a female sex determiner that is dominant over the ancestral sex determination system found in non-OB fish [20].
Taken together, the new insights obtained from the Nasonia genomes underscore the value of genomic studies of species with unusual biology. The demonstration that several interpretations previously given to explain peculiarities of the honey bee genome are unlikely also shows that caution is required when interpreting comparative results if data are available from only few studies. The increasing feasibility of sequencing multiple, well chosen genomes will no doubt open new opportunities to address a variety of interesting evolutionary questions.
References
-
Godfray H.C.J. Parasitoids: Behavioral and Evolutionary Ecology. Princeton University Press, Princeton 1994
-
Shuker D.M. West S.A. Information constraints and the precision of adaptation: sex ratio manipulation in wasps. Proc. Natl. Acad. Sci. USA. 2004; 101: 10363-10367
-
Parker Jr., E.D. Orzack S.H. Genetic variation for the sex ratio in Nasonia vitripennis. Genetics. 1985; 100: 93-105
-
West S.A. Sex Allocation. Princeton University Press, Princeton 2009
-
Werren J.H. Richards S. Desjardins C.A. Niehuis O. Gadau J. Colbourne J.K. Beukeboom L.W. Desplan C. Elsik C.G. Grimmelikhuijzen C.J. et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010; 327: 343-348
-
Robertson H.M. Gadau J. Wanner K.W. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol. Biol. 2010; 19: 121-136
-
de Graaf D.C. Aerts M. Brunain M. Desjardins C.A. Jacobs F.J. Werren J.H. Devreese B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010; 19: 11-26
-
Raychoudhury R. Grillenberger B.K. Gadau J. Bijlsma R. van de Zande L. Werren J.H. Beukeboom L.W. Phylogeography of Nasonia vitripennis (Hymenoptera) indicates a mitochondrial- Wolbachia sweep in North America. Heredity. 2010; (in press)
-
Hunt B.G. Wyder S. Elango N. Werren J.H. Zdobnov E.M. Yi S.V. Sociality is linked to rates of protein evolution in a highly social insect. Mol. Biol. Evol. 2010; (in press)
-
Linksvayer T.A. Wade M.J. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution. 2009; 63: 1685-1696
-
Toth A.L. Varala K. Newman T.C. Miguez F.E. Hutchison S.K. Willoughby D.A. Simons J.F. Egholm M. Hunt J.H. Hudson M.E. et al. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science. 2007; 318: 441-444
-
Wang J. Jemielity S. Uva P. Wurm Y. Gräff J. Keller L. An annotated cDNA library and microarray for large-scale gene-expression studies in the ant Solenopsis invicta. Genome Biol. 2007; 8: R9
-
Wurm Y. Uva P. Ricci F. Wang J. Jemielity S. Iseli C. Falquet L. Keller L. Fourmidable: a database for ant genomics. BMC Genomics. 2009; 10: 5
-
The Honey Bee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006; 443: 931-949
-
Wurm Y. Wang J. Keller L. Behavioral genomics: A, Bee, C, G, T. Curr. Biol. 2007; 17: R51-R53
-
Evans J.D. Aronstein K. Chen Y.P. Hetru C. Imler J.L. Jiang H. Kanost M. Thompson G.J. Zou Z. Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006; 15: 645-656
-
Desjardins C.A. Perfectti F. Bartos J.D. Enders L.S. Werren J.H. The genetic basis of interspecies host preference differences in the model parasitoid Nasonia. Heredity. 2010; (in press)
-
Loehlin D.W. Oliveira D.C. Edwards R. Giebel J.D. Clark M.E. Cattani M.V. van de Zande L. Verhulst E.C. Beukeboom L.W. Muñoz-Torres M. et al. Sex-specific wing size evolution by noncoding changes at doublesex in closely related species of Nasonia. PLoS Genet. 2010; 6: e1000821
-
Oliveira D.C.S.G. Werren J.H. Verhulst E.C. Giebel J.D. Kamping A. Beukeboom L.W. van de Zande L. Identification and characterization of the doublesex gene of Nasonia. Insect Mol. Biol. 2009; 18: 315-324
-
Roberts R.B. Ser J.R. Kocher T.D. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009; 326: 998-1001
Article Info
Identification
DOI: https://doi.org/10.1016/j.cub.2010.01.027
Copyright
© 2010 Elsevier Ltd. Published by Elsevier Inc.