Ants, Bees, Genomes & Evolution @ Queen Mary University London
Published: October 2017
The Global Ant Genomics Alliance (GAGA)
The Global Ant Genomics Alliance (GAGA) Consortium
Myrmecological News, October 2017, Volume 25, Pages 61-66
For centuries, scholars, the lay public, and children alike have been enthralled by ants because of their fascinating behaviors and social organization. Decades of biological research have documented that the ants have evolved a stunning global diversity (WARD 2014) (Fig. 1). More than 15,000 species belonging to over 330 genera have been named to date (ANTWEB 2017) and new species are discovered almost every day. While some of these species are extraordinarily rare, known from a single collection or site, others have spread around the world. Collectively, ants are among the most abundant insects in most terrestrial ecosystems on every continent except Antarctica. Over more than 100 million years of history, the ants have been impressive social innovators. They evolved advanced division of labor; assembly line processing of food resources; mass-raiding predatory columns; seed-harvesting and storage; sophisticated chemical, acoustic and visual communication systems; fungus-farming for food; herding aphids as livestock for meat and honeydew; practicing biological and chemical pest control; and social immune systems akin to efficient public health systems (HÖLLDOBLER & WILSON 1990, BOURKE & FRANKS 1995, CREMER & SIXT 2009). What sets ants apart from all but a handful of other animal lineages is their advanced reproductive division of labor with specialized, morphologically distinct castes for breeding (queens), nursing and food-gathering (workers), and sometimes defense (soldiers), all of which are unconditionally devoted to a single colony (CROZIER & PAMILO 1996). Dozens of ant species have been intensively studied by ecologists, geneticists, biogeographers, and evolutionary biologists, both in the field and in the laboratory. Some ant species have also become model organisms in studies of epidemiology, aging, development, and neurobiology. During the last seven years, 20 completely sequenced ant genomes have been published, so the ants are among the genomically best-known insect families (Tab. 1). These studies have provided remarkable information about the genetic underpinnings of key adaptations such as those that accompanied fungus farming and allowed communication and recognition systems to become highly sophisticated (SIMOLA & al. 2013), but the big comparative picture of genomic diversity across the entire family is still lacking. The fact that ant genomes tend to be modest in size has greatly facilitated genome sequencing efforts so far, and now allows us to pursue much more ambitious objectives.
We have recently launched the Global Ant Genomics Alliance (GAGA) to fundamentally advance the multidisciplinary genomics-based study of ant biology. Through this program, we will obtain full high-quality genome sequences of at least one representative of each ant genus that can be feasibly collected. GAGA will provide the overarching framework necessary for a series of large-scale comparative research programs to explain the genomic underpinnings of key innovations that globally characterize the ants as a group, as well as those that characterize the distinct subgroups.
Fig. 1
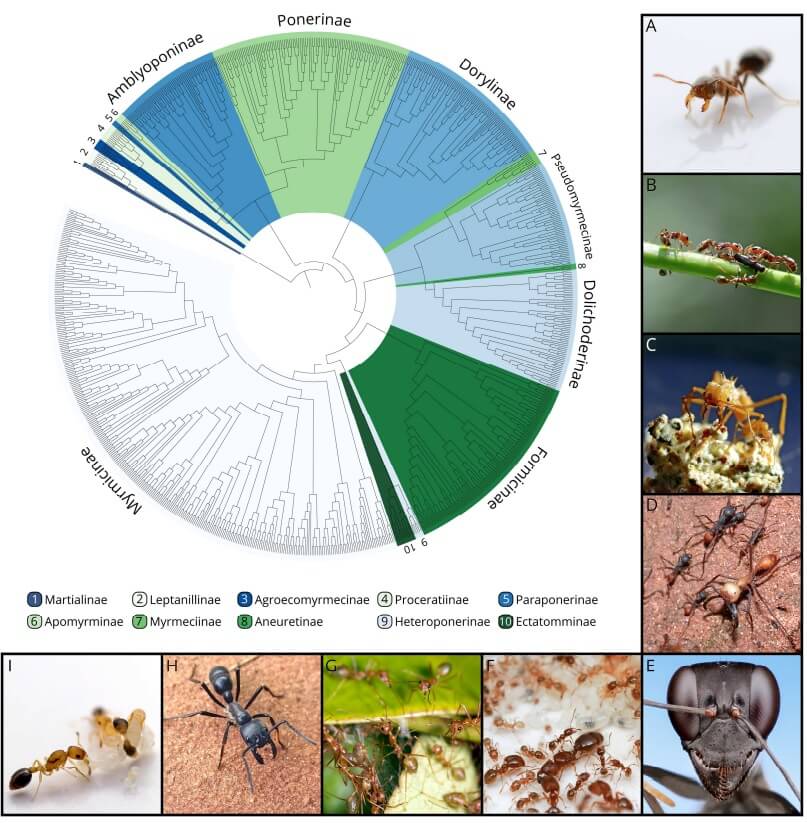
The evolutionary and biological diversity of the ants (Formicidae) captured with their tree of life and representative images. The phylogenetic tree is based on over 1000 species from 287 genera and shows the relationship between the seventeen currently recognized subfamilies (BRANSTETTER & al. 2017). (A) Threat display of a Solenopsis fire ant worker (Myrmicinae); (B) plant ants of the genus Pseudomyrmex (Pseudomyrmicinae) on their mutualistic Acacia host plant in Cerro Zuela (Panama); (C) a minor and a medium worker of the fungus-farming ant Acromyrmex echinatior (Myrmicinae) caring for their fungal symbiont; (D) a raiding column of the army ant Eciton burchellii (Dorylinae); (E) a Gigantiops desctructor worker (Formicinae); (F) a lab colony of Pheidole californica (Myrmicinae); (G) cooperation in nest construction between workers of Oecophylla smaragdina (Formicinae) in Queensland, Australia; (H) foraging worker of Dinoponera australis (Ponerinae); (I) broodcare behavior in a lab colony of Cardiocondyla obscurior (Myrmicinae). Photo credits: Lukas Schrader (A, B, C, G, I); Alex Wild (D, E, F, H).
At the same time, GAGA will actively encourage sequencing efforts that focus on more detailed species-level genomic comparisons in lineages (often tribes) of ants with unique life histories that are likely to have retained more specific signatures of molecular adaptation in their genomes. We expect such projects to be coordinated by PI groups with their own funding independent of the GAGA objectives, but we hope they will interact with GAGA to share data and facilitate comparisons of genomic data sets with sister lineages that will be sequenced under the auspices of GAGA.
Tab. 1
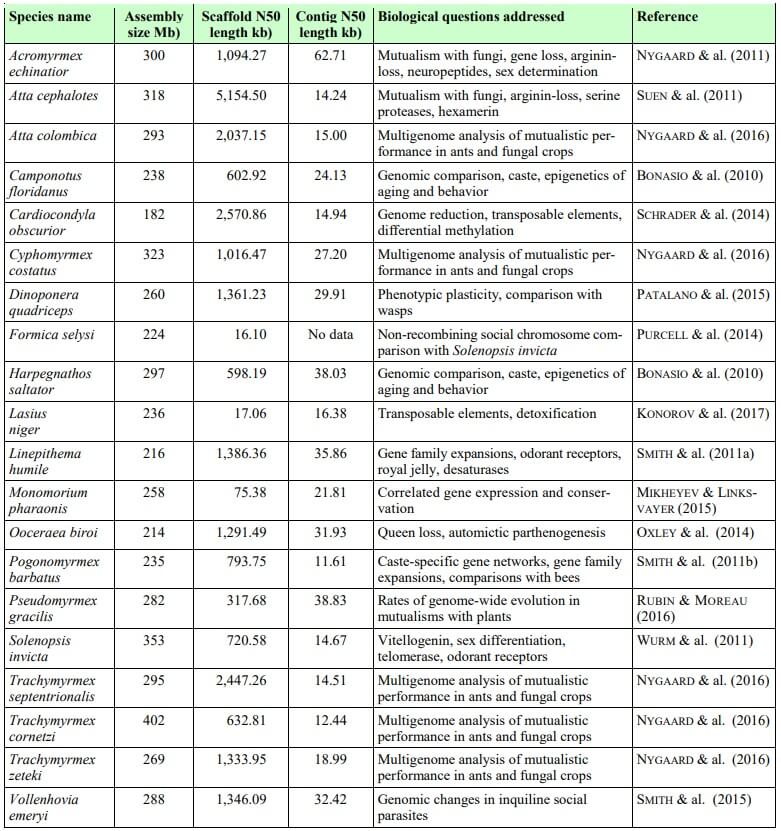
The 20 ant species for which sequenced genomes are available, in alphabetical order. Scaffold N50 and contig N50 are standard parameters for evaluating and comparing the quality of genome assemblies. Different sequencing strategies and assembly methods produce draft genomes of varying quality as illustrated by the figures in the table. The GAGA consortium aims to produce a consensus on the genome assembly standard for ant genomics research. With access to the best possible sequencing technologies, we expect to be able to produce top-quality assemblies with only a few thousand gaps, similar to the latest reference genomes produced for humans. We anticipate that GAGA will thus be able to address many biological questions that could not be resolved with existing fragmented and less well assembled genomes. Examples of such questions include chromosome evolution, synteny between genes far apart on chromosomes, reconstruction of gene-family evolutionary histories, and the evolution of gene regulatory regions.
GAGA is among the most ambitious new global initiatives for comparative genomics (PENNISI 2017), following in the footsteps of the recently completed avian comparative genomics project (ZHANG & al. 2014), which demonstrated the power of large-scale collaborations for uncovering global patterns of phenotypic diversification, evolutionary convergence, and genome-level evolutionary constraints. By applying this comprehensive approach to elucidate the global genomic diversity of the ants, we aim to address a series of fundamental questions in ecology, evolution, and developmental biology, and we expect to obtain significant novel insights. Because the ants are already one of the best-studied insect lineages, they offer many opportunities to combine genomic insights (NYGAARD & WURM 2015) with comparative data on natural history, adaptive radiation, ecological functionality, species distributions, and complex behavior coordinated by advanced communication. Other opportunities for significant improvements in biological understanding will emerge from the comparative analyses of genomic characteristics that we hope to connect with, for example, degrees of division of labor between queen and worker castes, and ancestral haplodiploid or evolutionarily derived novel sex determination systems.
In addition to the genomic data, we will coordinate the compilation of macroecological datasets on global ant biodiversity and biogeography to be supplemented by as much core life-history data as the global community can provide. Life histories of ant species and genera vary dramatically in terms of colony structure, reproduction, diet, habitat, range size, interactions with other ants, and ecological dominance. This extraordinary diversity offers unique opportunities for examining the molecular and genomic correlates of ecological abundance and life-history traits (DUNN & al. 2009, GIBB & al. 2015) and for assessing putative explanations for such correlations from first principles. We expect that GAGA will significantly advance our knowledge of phylogenomics, providing unprecedented resolution of the ant tree of life that will allow novel understanding of the forces that drove the adaptive radiations of the ant subfamilies. Explaining the evolution and elaboration of physically differentiated castes will be of paramount importance, both in terms of phenotypic functionality and developmental biology. Independent of phylogeny, GAGA will also offer key insights in the convergent evolution of adaptations to extreme environments and the recurrent origins of symbioses with microbes and macroscopic partners serving nutritional or protective purposes.
Finally, the ants stand out as the most diverse lineage for the expression of reproductively altruistic behavior, mediated by an enormous variation in primarily chemical communication systems. Although haplodiploidy has been retained throughout the ants, some lineages evolved parthenogenesis or experimented with mixtures of sexual and asexual propagation of either worker or queen castes (SCHWANDER & al. 2010). Haplodiploidy also endowed the ants with a unique burden of potential reproductive conflicts over sex-ratio investments, the regulation of male-production by workers, and the emergence of multiqueen colonies in most if not all major lineages (BOURKE & FRANKS 1995, CROZIER & PAMILO 1996, BOOMSMA & al. 2014). The investigation of genomic rearrangements that induce changes in social organization and control of potential reproductive conflicts has only just begun and stands to gain significantly from the large-scale availability of ant genomes (WANG & al. 2013, PURCELL & al. 2014). Funding for sequencing the first ca. 200 genomes has been secured via a Chinese biodiversity genomics grant embedded in the Kunming Institute of Zoology of the Chinese Academy of Sciences, and we anticipate that funding for 100 - 200 more genomes will become available as GAGA develops and reaches its full potential in the coming one to two years. With regard to technology, GAGA aims to produce high-quality genome assemblies via complementary approaches that can accommodate both common ant species, for which sufficient biomass is relatively easy to obtain, and rare species, for which genomes will have to be generated from minute quantities of high quality DNA.
GAGA will be supported by a worldwide bioinformatics analysis group with rich experience in comparative ant genomics. The consortium will further provide transparent guidelines for accessing the most cutting-edge sequencing technologies, so the best value for the money can be ensured throughout the GAGA program. We aim to establish a common standard for data collection and analysis, which will allow GAGA to serve the global community of ant biologists in the best possible way, and to make sure that GAGA-generated genome sequences will comply with the ambitious assembly and annotation standards that soon will be universally required for published reference genomes. We are reaching out to the international community of myrmecologists and other biological specialists to participate – with collecting efforts, unique expertise in the annotation of specific gene families, and with grant applications to fund complementary genome-sequencing efforts focusing on specific ant groups for which researchers aim to obtain multiple species-level genomes and for which GAGA can provide the outgroup genomes. We hope that the global community of ant biologists will spearhead new research programs on wide-ranging research questions that can be facilitated under the GAGA umbrella. GAGA will ensure fair credit to all contributing researchers independent of their type of employment and career stage. Collection trips to obtain the first ca. 100 ant species are planned for 2017 and 2018 and we hope to hear from other myrmecologists who are interested in joining the global sampling endeavor.
Interested researchers can contact GAGA via the project website (www.antgenomics.dk) to learn more about the initiative, to inspect and develop the list of ant species targeted for collection and sequencing, to propose new or alternative species within genera, or to offer expertise to contribute to ongoing projects. We are also eager to hear from PIs who would like to propose and fund larger comparative projects so they can spearhead analyses for complementary papers, similar to the process that generated a number of flagship papers in the bird program on which the coordinators were neither first nor last authors. We aim to publish the overarching results of the GAGA efforts as a coordinated set of papers in 2021. We anticipate that not all global ant genome-sequencing efforts during the coming five years can be embedded into GAGA, but we will be eager to discuss pre-publication data-sharing with other groups to encourage sharing of credits in the timeliest way possible.
In sum, we consider GAGA to be a platform for numerous existing research programs and a key instrument for facilitating novel collaborations that address fundamental questions about one of the most interesting and most amazing animal lineages on the planet – the ants.
References
-
ANTWEB 2017: AntWeb v6.74. – <https://www.antweb.org>, retrieved on 23 May 2017.
-
BONASIO, R., ZHANG, G., YE, C., MUTTI, N.S., FANG, X., QIN, N., DONAHUE, G., YANG, P., LI, Q., LI, C., ZHANG, P., HUANG, Z., BERGER, S.L., REINBERG, D., WANG, J. & LIEBIG, J. 2010: Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. – Science 329: 1068-1071.
-
BOOMSMA, J.J., HUSZÁR, D.B. & PEDERSEN, J.S. 2014: The evolution of multiqueen breeding in eusocial lineages with permanent physically differentiated castes. – Animal Behaviour 92: 214-251.
-
BOURKE, A.F.G. & FRANKS, N.R. 1995: Social evolution in ants. – Princeton University Press, Princeton, NJ, xiii, 529 pp. BRANSTETTER, M.G., LONGINO, J.T., WARD, P.S. & FAIRCLOTH, B.C. 2017: Enriching the ant tree of life: enhanced UCE bait set for genome-scale phylogenetics of ants and other Hymenoptera. – Methods in Ecology and Evolution, doi: 10.1111/ 2041-210X.12742.
-
CREMER, S. & SIXT, M. 2009: Analogies in the evolution of individual and social immunity. – Philosophical Transactions of the Royal Society B-Biological Sciences 364: 129-142.
-
CROZIER, R.H. & PAMILO, P. 1996: Evolution of social insect colonies.– Oxford University Press, Oxford, UK, 306 pp.
-
DUNN, R.R., AGOSTI, D., ANDERSEN, A.N., ARNAN, X., BRÜHL, C.A., CERDÁ, X., ELLISON, A.M., FISHER, B.L., FITZPATRICK, M.C., GIBB, H., GOTELLI, N.J., GOVE, A.D., GUENARD, B., JANDA, M., KASPARI, M., LAURENT, E.J., LESSARD, J.P., LONGINO, J.T., MAJER, J.D., MENKE, S.B., MCGLYNN, T.P., PARR, C.L., PHILPOTT, S.M., PFEIFFER, M., RETANA,J., SUAREZ, A.V., VASCONCELOS, H.L., WEISER, M.D. & SANDERS, N.J. 2009: Climatic drivers of hemispheric asymmetry in global patterns of ant species richness. – Ecology Letters 12: 324-333.
-
GIBB, H., SANDERS, N.J., DUNN, R.R., WATSON, S., PHOTAKIS, M., ABRIL, S., ANDERSEN, A.N., ANGULO, E., ARMBRECHT, I., ARNAN, X., BACCARO, F.B., BISHOP, T.R., BOULAY, R., CASTRACANI, C., DEL TORO, I., DELSINNE, T., DIAZ, M., DONOSO, D.A., ENRIQUEZ, M.L., FAYLE, T.M., FEENER, D.H., Jr., FITZPATRICK, M.C., GOMEZ, C., GRASSO, D.A., GROC, S., HETERICK, B., HOFFMANN, B.D., LACH, L., LATTKE, J., LEPONCE, M., LESSARD, J.P., LONGINO, J., LUCKY, A., MAJER, J., MENKE, S.B., MEZGER, D., MORI, A., MUNYAI, T.C., PAKNIA, O., PEARCE-DUVET, J., PFEIFFER, M., PHILPOTT, S.M., DE SOUZA, J.L., TISTA, M., VASCONCELOS, H.L., VONSHAK, M. & PARR, C.L. 2015: Climate mediates the effects of disturbance on ant assemblage structure. – Proceedings of the Royal Society BBiological Sciences 282: 20150418.
-
HÖLLDOBLER, B. & WILSON, E.O. 1990: The ants.– Belknap Press of Harvard University Press, Cambridge, MA, XII + 732 pp.
-
KONOROV, E.A., NIKITIN, M.A., MIKHAILOV, K.V., LYSENKOV, S.N., BELENKY, M., CHANG, P.L., NUZHDIN, S.V. & SCOBEYEVA, V.A. 2017: Genomic exaptation enables Lasius niger adaptation to urban environments. – BioMed Central Evolutionary Biology 17 (Supplement 1): art. 39.
-
MIKHEYEV, A.S. & LINKSVAYER, T.A. 2015: Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. – Elife 4: art. e04775.
-
NYGAARD, S., HU, H., LI, C., SCHIOTT, M., CHEN, Z., YANG, Z., XIE, Q., MA, C., DENG, Y., DIKOW, R.B., RABELING, C., NASH, D.R., WCISLO, W.T., BRADY, S.G., SCHULTZ, T.R., ZHANG, G. & BOOMSMA, J.J. 2016: Reciprocal genomic evolution in the ant-fungus agricultural symbiosis. – Nature Communications 7: art. 12233.
-
NYGAARD, S. & WURM, Y. 2015: Ant genomics (Hymenoptera: Formicidae): challenges to overcome and opportunities to seize. – Myrmecological News 21: 59-72.
-
NYGAARD, S., ZHANG, G., SCHIOTT, M., LI, C., WURM, Y., HU, H., ZHOU,J., JI, L., QIU, F., RASMUSSEN, M., PAN, H., HAUSER, F., KROGH, A., GRIMMELIKHUIJZEN, C.J., WANG, J. & BOOMSMA, J.J. 2011: The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. – Genome Research 21: 1339-1348.
-
OXLEY, P.R., JI, L., FETTER-PRUNEDA, I., MCKENZIE, S.K., LI, C., HU, H., ZHANG, G. & KRONAUER, D.J. 2014: The genome of the clonal raider ant Cerapachys biroi. – Current Biology 24: 451-458.
-
PATALANO, S., VLASOVA, A., WYATT, C., EWELS, P., CAMARA, F., FERREIRA, P.G., ASHER, C.L., JURKOWSKI, T.P., SEGONDSPICHON, A., BACHMAN, M., GONZALEZ-NAVARRETE, I., MINOCHE, A.E., KRUEGER, F., LOWY, E., MARCET-HOUBEN, M., RODRIGUEZ-ALES, J.L., NASCIMENTO, F.S., BALASUBRAMANIAN, S., GABALDON, T., TARVER, J.E., ANDREWS, S., HIMMELBAUER, H., HUGHES, W.O., GUIGO, R., REIK, W. & SUMNER, S. 2015: Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. – Proceedings of the National Academy of Sciences of the United States of America 112: 13970-13975.
-
PENNISI, E. 2017: Sequencing all life captivates biologists. – Science 355: 894-895.
-
PURCELL, J., BRELSFORD, A., WURM, Y., PERRIN, N. & CHAPUISAT, M. 2014: Convergent genetic architecture underlies social organization in ants. – Current Biology 24: 2728-2732.
-
RUBIN, B.E. & MOREAU, C.S. 2016: Comparative genomics reveals convergent rates of evolution in ant-plant mutualisms. – Nature Communications 7: art. 12679.
-
SCHRADER, L., KIM, J.W., ENCE, D., ZIMIN, A., KLEIN, A., WYSCHETZKI, K., WEICHSELGARTNER, T., KEMENA, C., STOKL, J., SCHULTNER, E., WURM, Y., SMITH, C.D., YANDELL, M., HEINZE, J., GADAU, J. & OETTLER, J. 2014: Transposable element islands facilitate adaptation to novel environments in an invasive species. – Nature Communications 5: art. 5495.
-
SCHWANDER, T., LO, N., BEEKMAN, M., OLDROYD, B.P. & KELLER, L. 2010: Nature versus nurture in social insect caste differentiation. – Trends in Ecology & Evolution 25: 275-282.
-
SIMOLA, D.F., WISSLER, L., DONAHUE, G., WATERHOUSE, R.M., HELMKAMPF, M., ROUX, J., NYGAARD, S., GLASTAD, K.M., HAGEN, D.E., VILJAKAINEN, L., REESE, J.T., HUNT, B.G., GRAUR, D., ELHAIK, E., KRIVENTSEVA, E.V., WEN, J., PARKER, B.J., CASH, E., PRIVMAN, E., CHILDERS, C.P., MUNOZTORRES, M.C., BOOMSMA, J.J., BORNBERG-BAUER, E., CURRIE, C.R., ELSIK, C.G., SUEN, G., GOODISMAN, M.A., KELLER, L., LIEBIG, J., RAWLS, A., REINBERG, D., SMITH, C.D., SMITH, C.R., TSUTSUI, N., WURM, Y., ZDOBNOV, E.M., BERGER, S.L. & GADAU, J. 2013: Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. – Genome Research 23: 1235-1247.
-
SMITH, C.D., ZIMIN, A., HOLT, C., ABOUHEIF, E., BENTON, R., CASH, E., CROSET, V., CURRIE, C.R., ELHAIK, E., ELSIK, C.G., FAVE, M.J., FERNANDES, V., GADAU, J., GIBSON, J.D., GRAUR, D., GRUBBS, K.J., HAGEN, D.E., HELMKAMPF, M., HOLLEY, J.A., HU, H., VINIEGRA, A.S., JOHNSON, B.R., JOHNSON, R.M., KHILA, A., KIM, J.W., LAIRD, J., MATHIS, K.A., MOELLER, J.A., MUNOZ-TORRES, M.C., MURPHY, M.C., NAKAMURA, R., NIGAM, S., OVERSON, R.P., PLACEK, J.E., RAJAKUMAR, R., REESE, J.T., ROBERTSON, H.M., SMITH, C.R., SUAREZ, A.V., SUEN, G., SUHR, E.L., TAO, S., TORRES, C.W., VAN WILGENBURG, E., VILJAKAINEN, L., WALDEN, K.K., WILD, A.L., YANDELL, M., YORKE, J.A. & TSUTSUI, N.D. 2011a: Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). – Proceedings of the National Academy of Sciences of the United States of America 108: 5673-5678.
-
SMITH, C.R., HELMS CAHAN, S., KEMENA, C., BRADY, S.G., YANG, W., BORNBERG-BAUER, E., ERIKSSON, T., GADAU, J., HELMKAMPF, M., GOTZEK, D., OKAMOTO MIYAKAWA, M., SUAREZ, A.V. & MIKHEYEV, A. 2015: How do genomes create novel phenotypes? Insights from the loss of the worker caste in ant social parasites. – Molecular Biology and Evolution 32: 2919-2931.
-
SMITH, C.R., SMITH, C.D., ROBERTSON, H.M., HELMKAMPF, M., ZIMIN, A., YANDELL, M., HOLT, C., HU, H., ABOUHEIF, E., BENTON, R., CASH, E., CROSET, V., CURRIE, C.R., ELHAIK, E., ELSIK, C.G., FAVE, M.J., FERNANDES, V., GIBSON, J.D., GRAUR, D., GRONENBERG, W., GRUBBS, K.J., HAGEN, D.E., VINIEGRA, A.S., JOHNSON, B.R., JOHNSON, R.M., KHILA, A., KIM, J.W., MATHIS, K.A., MUNOZ-TORRES, M.C., MURPHY, M.C., MUSTARD, J.A., NAKAMURA, R., NIEHUIS, O., NIGAM, S., OVERSON, R.P., PLACEK, J.E., RAJAKUMAR, R., REESE, J.T., SUEN, G., TAO, S., TORRES, C.W., TSUTSUI, N.D., VILJAKAINEN, L., WOLSCHIN, F. & GADAU, J. 2011b: Draft genome of the red harvester ant Pogonomyrmex barbatus. – Proceedings of the National Academy of Sciences of the United States of America 108: 5667-5672.
-
SUEN, G., TEILING, C., LI, L., HOLT, C., ABOUHEIF, E., BORNBERGBAUER, E., BOUFFARD, P., CALDERA, E.J., CASH, E., CAVANAUGH, A., DENAS, O., ELHAIK, E., FAVE, M.J., GADAU, J., GIBSON, J.D., GRAUR, D., GRUBBS, K.J., HAGEN, D.E., HARKINS, T.T., HELMKAMPF, M., HU, H., JOHNSON, B.R., KIM, J., MARSH, S.E., MOELLER, J.A., MUNOZ-TORRES, M.C., MURPHY, M.C., NAUGHTON, M.C., NIGAM, S., OVERSON, R., RAJAKUMAR, R., REESE, J.T., SCOTT, J.J., SMITH, C.R., TAO, S., TSUTSUI, N.D., VILJAKAINEN, L., WISSLER, L., YANDELL, M.D., ZIMMER, F., TAYLOR, J., SLATER, S.C., CLIFTON, S.W., WARREN, W.C., ELSIK, C.G., SMITH, C.D., WEINSTOCK, G.M., GERARDO, N.M. & CURRIE, C.R. 2011: The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. – Public Library of Science Genetics 7: art. e1002007.
-
WANG, J., WURM, Y., NIPITWATTANAPHON, M., RIBA-GROGNUZ, O., HUANG, Y.C., SHOEMAKER, D. & KELLER, L. 2013: A Y-like social chromosome causes alternative colony organization in fire ants. – Nature 493: 664-668.
-
WARD, P.S. 2014: The phylogeny and evolution of ants. – Annual Review of Ecology, Evolution, and Systematics 45: 23-43.
-
WURM, Y., WANG,J., RIBA-GROGNUZ, O., CORONA, M., NYGAARD, S., HUNT, B.G., INGRAM, K.K., FALQUET, L., NIPITWATTANAPHON, M., GOTZEK, D., DIJKSTRA, M.B., OETTLER, J., COMTESSE, F., SHIH, C.J., WU, W.J., YANG, C.C., THOMAS, J., BEAUDOING, E., PRADERVAND, S., FLEGEL, V., COOK, E.D., FABBRETTI, R., STOCKINGER, H., LONG, L., FARMERIE, W.G., OAKEY, J., BOOMSMA, J.J., PAMILO, P., YI, S.V., HEINZE, J., GOODISMAN, M.A., FARINELLI, L., HARSHMAN, K., HULO, N., CERUTTI, L., XENARIOS, I., SHOEMAKER, D. & KELLER, L. 2011: The genome of the fire ant Solenopsis invicta. – Proceedings of the National Academy of Sciences of the United States of America 108: 5679-5684.
-
ZHANG, G., JARVIS, E.D. & GILBERT, M.T. 2014: Avian genomes. A flock of genomes. Introduction. – Science 346: 1308-1309.