Ants, Bees, Genomes & Evolution @ Queen Mary University London
Published: 24 January 2022
The era of reference genomes in conservation genomics
European Reference Genome Atlas (ERGA) Consortium
Trends in Ecology & Evolution Volume 37, Issue 3, March 2022, Pages 197-202
Abstract
Progress in genome sequencing now enables the large-scale generation of reference genomes. Various international initiatives aim to generate reference genomes representing global biodiversity. These genomes provide unique insights into genomic diversity and architecture, thereby enabling comprehensive analyses of population and functional genomics, and are expected to revolutionize conservation genomics.
Conservation, genomics, and reference genomes
In 2020 both the United Nations Biodiversity Summit and the European Environment Agency emphasized the accelerating global loss of biodiversity (https://www.un.org/pga/75/united-nations-summit-on-biodiversity/; https://www.eea.europa.eu/highlights/latest-evaluation-shows-europes-nature). We are in the sixth mass extinction. Although the primary route to preserving biodiversity comprises protection of species and restoration of habitats and ecosystems, genomics provides a rapidly expanding array of novel tools to characterize biodiversity and assist such conservation efforts. The need for immediate actions that help to reverse the current biodiversity decline has prompted national and international initiatives aimed at expanding the genomic reference resources available for biodiversity research and conservation across the tree of life (Box 1). Many of these efforts collectively contribute to the Earth BioGenome Project (EBP) that aims to catalog and characterize the genomes of all of Earth’s eukaryotic biodiversity. A large and inclusive community of scientists has recently gathered as the European hub of the EBP to promote the generation of a European Reference Genome Atlas (ERGA; www.erga-biodiversity.eu). This initiative is building a pan-European open access infrastructure to streamline ethical and legally compliant sample and metadata collection [1.], sequencing and assembly (see Glossary) [2.], annotation [3.], and release in public archives of high-quality genomic information, thus creating reference genomes for a wide variety of eukaryotic species (Box 1).
Box 1
Sequencing the tree of life
International initiatives aimed at generating genomic resources, and particularly reference genomes, have flourished in recent years. Some focus on specific taxa, such as the Vertebrate Genomes Project, Bird Genome 10K Project, Bat1K Project, Global Invertebrate Genomics Alliance, 10 000 Plant Genomes Project, and 1000 Fungal Genomes project. Others focus on geographic regions, such as the California Conservation Genomics Project, Darwin Tree of Life for Britain and Ireland, Catalan Initiative for the Earth BioGenome Project in the Catalan territories, Endemixit in Italy, Norwegian Earth Biogenome Project, and SciLifeLab in Sweden, on applications such as the LOEWE Translational Biodiversity Genomics in Germany, or on ecological systems such as the Aquatic Symbiosis Genomics project. Collectively part of the Earth BioGenome Project (EBP), in Europe these initiatives are organized under the umbrella of the European Reference Genome Atlas (ERGA).
A genome atlas of European biodiversity
ERGA is a pan-European scientific response to the current threats to biodiversity. Approximately one fifth of the ~200 000 eukaryotic species present in Europe can be inferred to be at risk of extinction according to the International Union for Conservation of Nature (IUCN) Red List classification (this estimate only considers the assessed species; https://www.iucn.org/regions/europe/our-work/biodiversity-conservation/european-red-list-threatened-species).
ERGA aims to generate reference genomes of European eukaryotic species across the tree of life, including threatened, endemic, and keystone species, as well as pests and species important to agriculture, fisheries, and ecosystem function and stability. ERGA builds upon current genomic consortia in EU member states, EU Associated Countries, representatives of other countries within the European bioregion, and international collaborators. These reference genomes will address fundamental and applied questions in conservation, biology, and health. ERGA seeks to alert the EU about the potential of conservation genomics, and particularly the role of reference genomes, in biodiversity assessment, conservation strategies, and restoration efforts.
Reference genomes, by which we mean highly contiguous, accurate, and annotated genome assemblies, greatly enhance genomic studies, both experimentally and analytically [2.,4.]. A reference genome is a point representation of the structure and organization of the genome of a species. Similarly to type specimens in taxonomy, reference genomes serve as the standard for subsequent genomic studies [5.]. To cost-efficiently unravel the genomic diversity of species, multiple conspecific individuals can be resequenced and aligned to available reference genomes instead of being assembled de novo. Thus, reference genomes provide a comprehensive and fundamental framework onto which genomic variation can be mapped to characterize and ultimately aid in preserving genetic diversity [4.]. To this end, special attention should be paid to the origin of the individuals used as the reference because, if these are excessively divergent from the populations under study, this could compromise subsequent analyses. To overcome this issue, multiple conspecific genomes [6.] can now be summarized in the pangenome of a species [7.].
Until recently reference genomes have only been available for a handful of model organisms. Thanks to the consolidated and standardized efforts of international genome initiatives, the situation is rapidly changing. Recent technological advances provide a general strategy for generating chromosome-scale reference genomes for all organisms across the tree of life [2.]. These advances rely on a combination of single-molecule long-read sequencing [either PacBio Single Molecule Real-Time (SMRT) sequencing or Oxford Nanopore Technologies (ONT) sequencing] and/or linked reads [(e.g., transposase enzyme linked long-read sequencing (TELL-seq) or single-tube long fragment read (stLFR) sequencing] for contig assembly, optical mapping, and/or proximity ligation followed by high-throughput sequencing (Hi-C) for scaffolding [2.].
Decreasing costs, improved scalability, and increasing quality of sequencing technologies, combined with better algorithms and advances in computational power [2.], facilitate the establishment of reference genomes across the full spectrum of biodiversity. Importantly, reference genomes are fundamental for a comprehensive and accurate characterization of genomic information, for instance of structural features that cannot be inferred from fragmented genomes or reduced-representation sequencing approaches (Figure 1). Therefore, reference genomes coupled with resequencing data should become a standard in conservation genomics, facilitated by constantly evolving analytical methods.
Figure.1
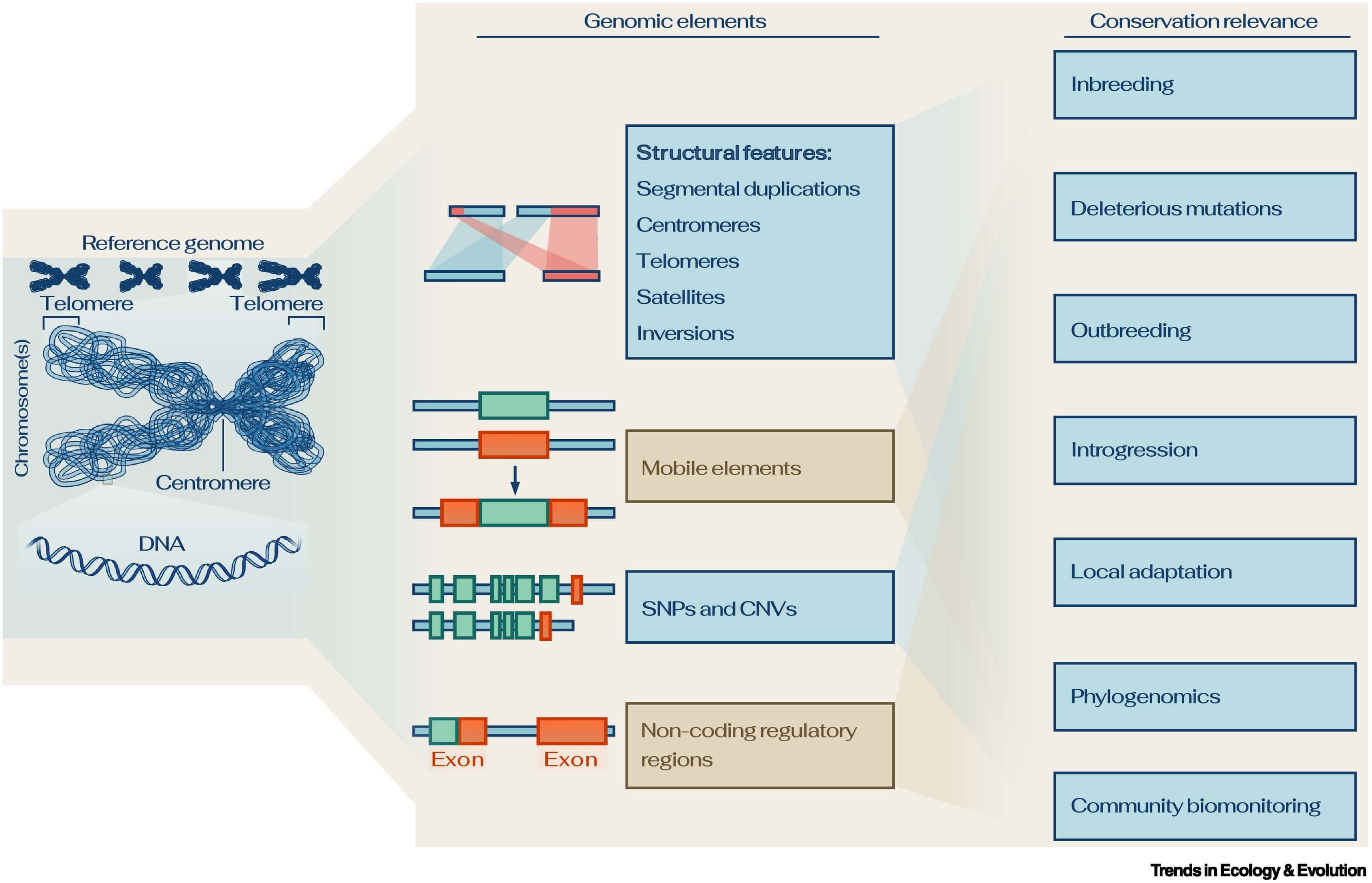
Reference genomes offer an (almost) complete record of the genome of a species.
They characterize genomic information more thoroughly than fragmented genomes can. Importantly, they reveal structural features which often remain elusive in fragmented genome sequences. These features are relevant for conservation genomics applications. Abbreviations: CNV, copy number variants; SNP, single nucleotide polymorphism.
Key contributions of reference genomes in conservation genomics
The full spectrum of genomic diversity
Reference genomes provide a view of the architecture of the genome, comprising both genic and intergenic regions. These include repetitive regions, some of which are challenging to assemble, such as segmental duplications, centromeres and telomeres, satellites, and mobile elements. Population genomics guided by reference genomes aids the identification of classical genetic variants, such as SNPs and copy number variants (CNVs), as well as structural variants that are particularly difficult to detect in fragmented and incomplete reference genomes alone, but are potentially important in adaptation to environmental change [8.].
Inbreeding and deleterious mutations
Assessments of inbreeding have long informed conservation and breeding programs, guiding genetic crosses and translocations of individuals. Although often estimated from a few loci, understanding the genetic architecture and accurately quantifying inbreeding and inbreeding depression require a genome-wide perspective, encompassing for example the number of genes involved, the presence of alleles with large effects, the role of deleterious recessive alleles, and heterozygote advantage [9.]. Although several questions remain, multiple studies have showcased the power of population genomics guided by reference genomes to identify runs of homozygosity as a means to estimate inbreeding, as well as to reveal the dynamics and fate of deleterious variation in threatened species (e.g., [10.]).
Outbreeding and introgression
Mating between individuals from genetically distinct lineages may lead to outbreeding depression due to chromosomal or genic incompatibilities, epistatic interactions, disruption of interactions between co-adapted genes, or the introduction of maladaptive variants into local populations. Population genomics guided by reference genomes greatly aids the disentanglement of these phenomena [11.]. Hybridization is a common evolutionary process that, through introgression, can promote the spread of adaptive variation and speciation. Anthropogenic hybridization and introgression, however, can be major threats to biodiversity and evolutionary heritage. Reference genomes facilitate the characterization of introgression patterns and dynamics as well as of admixture proportions, particularly of introgressed tracts along individual genomes [12.].
Local adaptation and genetic rescue
The use of reference genomes in population genomics facilitates the identification of traits under natural selection that form the basis and architecture of local adaptations, and ultimately of speciation. Reference genomes provide the functional and genomic contexts for regions influenced by selection, thereby enabling association of such loci with phenotypes important to adaptation and resilience. Identifying locally adapted variants can inform definitions of conservation units and identify optimal source populations for translocations to support genetic rescue [13.].
Phylogenetic diversity and phylogenomics
Phylogenetic diversity is essential for ecosystem stability and resilience, and is used to delineate evolutionarily distinct components of biodiversity to guide conservation priorities [e.g., evolutionary distinct and globally endangered (EDGE) species] [14.]. Genome-scale analyses based on hundreds or thousands of loci have become the gold standard for phylogenetic inference by capturing the evolutionary histories of the targeted taxa. Reference genomes serve as the basis for phylogenomic analyses because they greatly improve orthology inference at the DNA and protein levels, while also facilitating inferences based on genome organization.
Structure and function of communities
Reference genomes are particularly important in metagenomics and metatranscriptomics where total DNA, or complementary DNA (cDNA) derived from RNA, from entire communities is sequenced to understand community composition, abundance, function, and dynamics. Facilitated by the availability of reference genomes, metagenomics and metatranscriptomics have been mostly applied to microbial community samples. Eukaryotic reference genomes allow DNA/cDNA reads to be assigned to higher taxa within environmental samples, leading to a more complete characterization of communities from environmental DNA (eDNA) and RNA (eRNA). This approach represents a novel means to track changes in the composition, structure, and functioning of eukaryotic communities, and thus support the biomonitoring and management of taxonomic and functional diversity in entire ecosystems.
A collective effort to conserve biodiversity
Conservation efforts need to account for genomic diversity to optimize management strategies. Accounting for genomic diversity will aid in maintaining population viability and preserving adaptive potential to respond to environmental change. The availability of reference genomes will provide a solid, quantitative, and comparable foundation for biodiversity assessments, conservation, management, and restoration.
Author contributions
Giulio Formenti, Kathrin Theissinger, and Carlos Fernandes led the writing of the manuscript. Details on contributions to the initial discussion, to literature survey, drafting, reviewing of the manuscript and design of the figure can be found in the Supplemental information online.
Acknowledgments
We thank Fabien Condamine, Love Dalén, Richard Durbin, Bruno Fosso, Roderic Guigó, Marc Hanikenne, Alberto Pallavicini, Olga Vinnere Pettersson, Xavier Turon, and Detlef Weigel for their contributions to the manuscript, as well as the whole ERGA community for their ongoing support. We refer to the supplemental information online for acknowledgements for individual authors.
Declaration of interests
The authors declare no conflicts of interest.
Supplemental information
Table S1: Complete author list, including author contributions and affiliations.
mmc1.xlsx – [.XLSX, .04 MB]
References
-
Shaw F. et al. COPO: a metadata platform for brokering FAIR data in the life sciences. F1000Res. 2020; 9: 495
-
Rhie A. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021; 592: 737-746
-
Howe K.L. et al. Ensembl 2021. Nucleic Acids Res. 2021; 49: D884-D891
-
Brandies P. et al. The value of reference genomes in the conservation of threatened species. Genes (Basel). 2019; 10: 846
-
Ballouz S. et al. Is it time to change the reference genome?. Genome Biol. 2019; 20: 159
-
Valiente-Mullor C. et al. One is not enough: on the effects of reference genome for the mapping and subsequent analyses of short-reads. PLoS Comput. Biol. 2021; 17e1008678
-
Llamas B. et al. A strategy for building and using a human reference pangenome. F1000Res. 2019; 8: 1751
-
Mérot C. et al. A roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol. Evol. 2020; 35: 561-572
-
Kardos M. et al. Genomics advances the study of inbreeding depression in the wild. Evol. Appl. 2016; 9: 1205-1218
-
Dussex N. et al. Population genomics of the critically endangered kākāpō. Cell Genomics. 2021; 1100002
-
Leitwein M. et al. Associative overdominance and negative epistasis shape genome-wide ancestry landscape in supplemented fish populations. Genes (Basel). 2021; 12: 524
-
Rogers J. et al. The comparative genomics and complex population history of Papio baboons. Sci. Adv. 2019; 5eaau6947
-
Flanagan S.P. et al. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol. Appl. 2018; 11: 1035-1052
-
Owen N.R. et al. Global conservation of phylogenetic diversity captures more than just functional diversity. Nat. Commun. 2019; 10: 859
Glossary
Assembly, genome
a chromosome-level contiguous sequence of all chromosomes reconstructed using whole-genome sequencing reads, often aided by genetic maps or other information.
Evolutionary distinct and globally endangered (EDGE) species
species of high conservation priority.
Genetic rescue
a mitigation strategy for restoring intraspecific genetic diversity and reducing extinction risks in small, isolated, or inbred populations through induced gene flow.
Heterozygote advantage
when a heterozygous genotype has a higher relative fitness compared to a homozygous dominant or homozygous recessive genotype.
Hybridization
interbreeding of individuals from genetically distinct lineages.
Inbreeding depression
reduced fitness in offspring as a result of inbreeding – mating between closely related individuals.
Introgression
gene flow between hybridizing populations or species by backcrossing hybrids with one or both parental populations.
Metagenomics and metatranscriptomics
sequencing of DNA or RNA-derived cDNA extracted from environmental and bulk samples.
Outbreeding depression
reduced fitness in offspring from mating between genetically divergent individuals.
Pangenome
the entire set of DNA sequences (or genes) of a species represented by the core genome and the accessory genome.
Phylogenomics
the inference of the phylogenetic relationships among different lineages of organisms from genome-wide data.
Reference genome
a contiguous and accurate genome assembly representative of a species in which the coordinates of genes and other important features are annotated. Current definitions of reference genome quality are given in [2.] and https://www.earthbiogenome.org/assembly-standards.
Article Info
Publication History
Published online: January 24, 2022
Identification
DOI: https://doi.org/10.1016/j.tree.2021.11.008
Copyright
© 2021 Published by Elsevier Ltd.
User License
Creative Commons Attribution IGO (CC BY 3.0 IGO)
ScienceDirect
Access this article on ScienceDirect