Ants, Bees, Genomes & Evolution @ Queen Mary University London
Published: 07 May 2013
Duplication and concerted evolution in a master sex determiner under balancing selection
Eyal Privman, Yannick Wurm and Laurent Keller
Proceedings of the Royal Society B: Biological Sciences, 2013, 280:20122968
Abstract
The transformer (tra) gene is a key regulator in the signalling hierarchy controlling all aspects of somatic sexual differentiation in Drosophila and other insects. Here, we show that six of the seven sequenced ants have two copies of tra. Surprisingly, the two paralogues are always more similar within species than among species. Comparative sequence analyses indicate that this pattern is owing to the ongoing concerted evolution after an ancestral duplication rather than independent duplications in each of the six species. In particular, there was strong support for inter-locus recombination between the paralogues of the ant Atta cephalotes. In the five species where the location of paralogues is known, they are adjacent to each other in four cases and separated by only few genes in the fifth case. Because there have been extensive genomic rearrangements in these lineages, this suggests selection acting to conserve their synteny. In three species, we also find a signature of positive selection in one of the paralogues. In three bee species where information is available, the tra gene is also duplicated, the copies are adjacent and in at least one species there was recombination between paralogues. These results suggest that concerted evolution plays an adaptive role in the evolution of this gene family.
1. Introduction
In insects, tra has been found to be a conserved upstream component inducing female development by regulating sex-specific alternative splicing of downstream genes such as doublesex [1,2]. However, the primary signal activating tra varies among species. In some species, tra activation is controlled by a sex chromosome system, whereas in others (especially the 200 000 species of the order Hymenoptera that include the ants, bees, wasps and sawflies) there is a haplodiploid sex determination system where diploids develop into females and haploids into males [3]. Many hymenopterans use the complementary sex determination (CSD) mechanism, of which the honeybee Apis mellifera is the prototypical example. Heterozygosity at a single locus (the CSD locus) triggers female development in diploid individuals, while haploid individuals have a hemizygous CSD genotype and thus develop into males [4]. Likewise, homozygous diploid individuals have one allele and thus develop into diploid males, but these are generally unviable or sterile [5]. Selection against diploid males should thus favour rare alleles at the CSD locus by balancing selection [6].
The CSD locus was molecularly identified in the honeybee Ap. mellifera and found to be a homologue of tra [7]. In this species, the tra locus is duplicated [8]. One copy, named csd, is the primary signal of the sex determination pathway. It activates the second copy, named feminizer (fem), which is more conserved and retains the ancestral function of regulating doublesex and the downstream factors in the signal transduction cascade. Following the duplication csd was subject to positive selection, consistent with neofunctionalization of this paralogue. Two paralogues of tra were also found in the two other species of the genus Apis (Apis cerana and Apis dorsata). A gene tree, including these six sequences and a single homologue in the sister genus Bombus supported the view of a duplication in the Apis lineage, after the split from Bombus [8]. The finding of many highly diverged csd alleles in the three honeybee species is consistent with balancing selection acting on this locus to reduce production of diploid males [8].
The recent sequencing of seven ant genomes [9–14] provides an opportunity to investigate the gene duplication, sequence evolution, and selective forces acting on these genes in Hymenoptera. We will use the names csd and fem for the two paralogues in honeybees, but we will name them traA and traB in ants because their functional roles are still unknown. We chose these names rather than tra1 and tra2 because the name tra2 is already in use for a more distant paralogue that exists in many insects, including flies [15] and ants (data not shown).
2. Material and methods
(a) Identification of tra homologues
Homologues were identified in the seven ant genome assemblies [9–14] using translated BLAST [16] with the protein sequence of Acromyrmex echinatior tra [10] as the query. For Solenopsis invicta, we used an improved assembly of the genome (O. Riba-Grognuz 2012, unpublished data). Gene models were constructed using Maker [17] given as evidence homologous tra protein sequences and assembled transcriptome sequences for each species. Maker gene models were manually corrected using Apollo [18]. Coding sequences and predicted protein sequences are included in the electronic supplementary material.
(b) Previously published tra homologues
Genbank accession numbers for sequences used in phylogenetic and recombination analyses (amino acid, nucleotide sequences): Ap. mellifera fem (AAS86667, NM_001134828) and csd (AAS86653, NM_001011569); Ap. dorsata fem (ABV56232, EU100939) and csd (ABW36165, EU100933); Ap. cerana fem (ABV56230, EU100937) and csd (ABV58877, EU100908); Bombus terrestris fem (XP_003402358, XM_003402310); Nasonia vitripennis tra (NP_001128299).
Apis csd alleles used for recombination tests: Ap. mellifera: NM_001011569, AY350615, AY350617, AY350618, AY350616, EU100898, EU100896, EU100894, EU100892, EU100890, EU100888, EU100886, EU100899, EU100897, EU100895, EU100893, EU100891, EU100889, EU100887, EU100885, AY569720, AY569716, AY569712, AY569710, AY569708, AY569706, AY569704, AY569700, AY569698, AY569696, AY569694, AY569721, AY569717, AY569709, AY569707, AY569705, AY569703, AY569701, AY569699, AY569697, AY569695, AY352276, EU101390; Ap. cerana: EU100916, EU100914, EU100912, EU100910, EU100908, EU100906, EU100904, EU100902, EU100900, EU100915, EU100913, EU100911, EU100909, EU100907, EU100905, EU100903, EU100901; Ap. dorsata: EU100935, EU100933, EU100931, EU100929, EU100927, EU100925, EU100923, EU100921, EU100919, EU100917, EU100934, EU100932, EU100930, EU100928, EU100926, EU100924, EU100922, EU100920, EU100918.
(c) Alignment and phylogeny reconstruction
Protein sequences were aligned and the gene tree reconstructed using the simultaneous alignment and phylogeny Bayesian reconstruction algorithm implemented in BAli-Phy [19], with the LG + gamma substitution model, indel model RS07. Eight BAli-Phy chains were run in parallel until convergence. The posterior decoding alignment was used for all subsequent analyses. Unreliably aligned residues (approximately unbiased score less than 80%) were masked by replacing them with missing data characters (‘X’). The alignment was mapped back to the coding sequences to produce the corresponding codon sequence alignment (corresponding codons masked as ‘NNN’). The majority consensus tree was constructed (including splits with posterior probability greater than 50%). The maximum a posteriori tree was used for downstream analyses that require a fully bifurcating tree (recombination and positive selection tests).
(d) Positive selection and rate shift tests
The branch-site test for positive selection [20] implemented in PAML [21] was run on the masked BAli-Phy codon alignment to test each branch of the BAli-Phy maximum a posteriori tree for a dN/dS ratio (the omega parameter) greater than one. The null model (fixed omega of one) and the alternative model (free omega greater than one) were optimized for each branch and used to calculate the likelihood ratio test (LRT). The LRT confidence scores were adjusted to control the false discovery rate (FDR) using the Benjamini–Hochberg method [22]. A posterior probability of greater than 95% for a dN/dS ratio (the omega parameter) greater than 1 was considered significant evidence for positive selection in specific codon sites.
To test for acceleration of the evolutionary rate following duplication, we used PAML’s clade model C (six free parameters), which we ran once for each branch as the foreground branch. The C model allows the foreground branch to have a different rate than the rest of the tree for the third rate category [23]. We used M3 as the null model (five free parameters), because it allows three discrete rate categories similar to the C model, but no rate variation across the tree [24]. For each terminal branch, we calculated the LRT comparing the two models. We used the χ2-squared distribution with 1 d.f. to check for statistical significance.
(e) Recombination tests
Phylogenetic ‘splits networks’ were reconstructed based on evolutionary distances using the NeighborNet algorithm [25] implemented in the SplitsTree package [26]. Networks were visualized using the ‘equal angle’ algorithm [27]. The following tests were performed on the masked DNA alignments using the RDP4 package [28]: RDP [29] with internal and external reference, Maxchi [30], Geneconv [31], Bootscan [32] with neighbour joining trees, Chimaera [33], Siscan [34], 3seq [35], Lard [36] and Phylpro [37]. We used RDP4 to look for statistical support for specific recombination events. We applied different methods to datasets ranging in their sequence divergence: protein sequences from the full dataset, including the wasp and bee outgroups (18 sequences), coding DNA sequences from all seven ant species (13 sequences), from the five more closely related ant species (nine sequences), the four more closely related species (seven sequences) and the two most closely related species (Ac. echinatior and Atta cephalotes), together with S. invicta traA as an outgroup (four sequences).
We similarly analysed the honeybee tra homologues (fem and csd) from Ap. mellifera, Ap. cerana and Ap. dorsata. There are 43, 17 and 19 published csd allele sequences in Ap. mellifera, Ap. cerana and Ap. dorsata, respectively. To test for recombination between the two paralogues, we analysed sequence alignments that include the fem sequence and one csd allele sequence from each Apis species, and the B. terrestris fem as the outgroup (seven sequences). The sequences were codon-aligned and unreliably aligned codons were masked as ‘NNN’ using PRANK [38] and Guidance [39,40]. To test different csd alleles, we repeated these analyses 100 times, every time with a different random choice of one csd allele per species. We applied the aforementioned recombination tests to each of the 100 sequence alignments.
We used the ‘alignment uncertainty (AU) test of phylogenetic tree selection’ [41] to further confirm inter-locus recombination events that were inferred by the above tests in the ant and bee sequence alignments. Alignments were split into putative recombinant and non-recombinant regions according to the recombination points inferred by RDP4. Maximum-likelihood (ML) trees were constructed for each sub-alignment using PhyML v. 3.0.1beta [42] with the HKY85 substitution model, ML base frequencies, NNI and SPR tree search. Per position likelihood scores were calculated using baseml of the PAML package [21] for the two sub-alignment with each tree topology. Concel [43] was used to calculate the AU test to compare the ML tree topology of each sub-alignment with the tree from the other sub-alignment. p-values for multiple tests were adjusted to control the FDR using the Benjamini–Hochberg method [22].
(f) Conservation of synteny
Because we found that the two tra paralogues were adjacent in four of the seven genomes (At. cephalotes, S. invicta, Pogonomyrmex barbatus and Camponotus floridanus), we selected all genes having orthologues present in all four species, and determined the likelihood that a given pair of genes would be adjacent in these four species. This analysis was restricted to pairs of genes that were less than 50 000 bp apart, as are the tra paralogue pairs. A total of 56 156 pairs of such genes were found to be adjacent in one or more of the four genomes (see the electronic supplementary material, table S1). We then determined the number of cases where a given pair of genes was adjacent in all four genomes to obtain a probability of synteny being conserved in the four species studied.
3. Results
(a) Concerted evolution of tra paralogues
Two copies of the tra gene were found in six out of the seven sequenced ant genomes ([9,12], table 1) while the seventh genome, the leafcutter ant species Ac. echinatior, contained only one copy [10]. Surprisingly, a gene tree based on these tra sequences revealed greater similarity between the two paralogues in each of the six species than between any pair of orthologues across ant lineages (figure 1). This is the expected pattern if independent duplications occurred in each of the six species. However, it seems unlikely that a duplication of the same gene independently occurred in six of the seven lineages for which data are available. An alternative explanation is that the greater within- than between-species similarity is owing to concerted evolution. That is, inter-locus recombination and gene conversion events between the paralogues homogenize their sequences and increase their similarity [44].
Table 1 – Duplicated tra homologues found in the sequenced ant genomes.
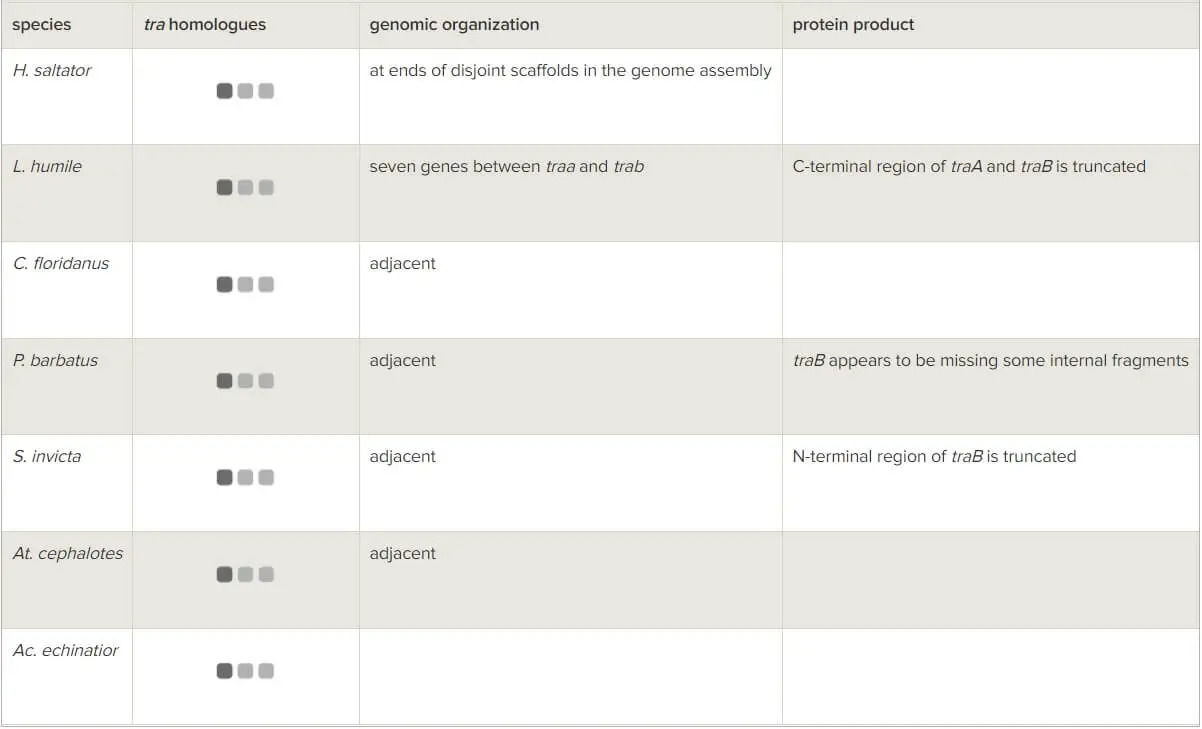
Figure 1
Gene tree of tra homologues in Hymenoptera. Stars indicate putative independent gene duplication events. Phylogeny was reconstructed using BAli-Phy from predicted protein sequences. Posterior probability support is indicated for each branch. Forward green arrows indicate significant acceleration of the evolutionary rate, and plus signs indicate significant evidence for positive diversifying selection. Backward blue arrow indicates significantly reduced rate (detailed results presented in tables 3 and 4). Species name abbreviations: nvit, Nasonia vitripennis; amel, Apis mellifera; acer, Apis cerana; ador, Apis dorsata; hsal, Harpegnathos saltator; lhum, Linepithema humile; cflo, Camponotus floridanus; pbar, Pogonomyrmex barbatus; sinv, Solenopsis invicta; aech, Acromyrmex echinatior; acep, Atta cephalotes. Gene name abbreviations: tra, transformer; fem, feminzer; csd, complementary sex determiner.
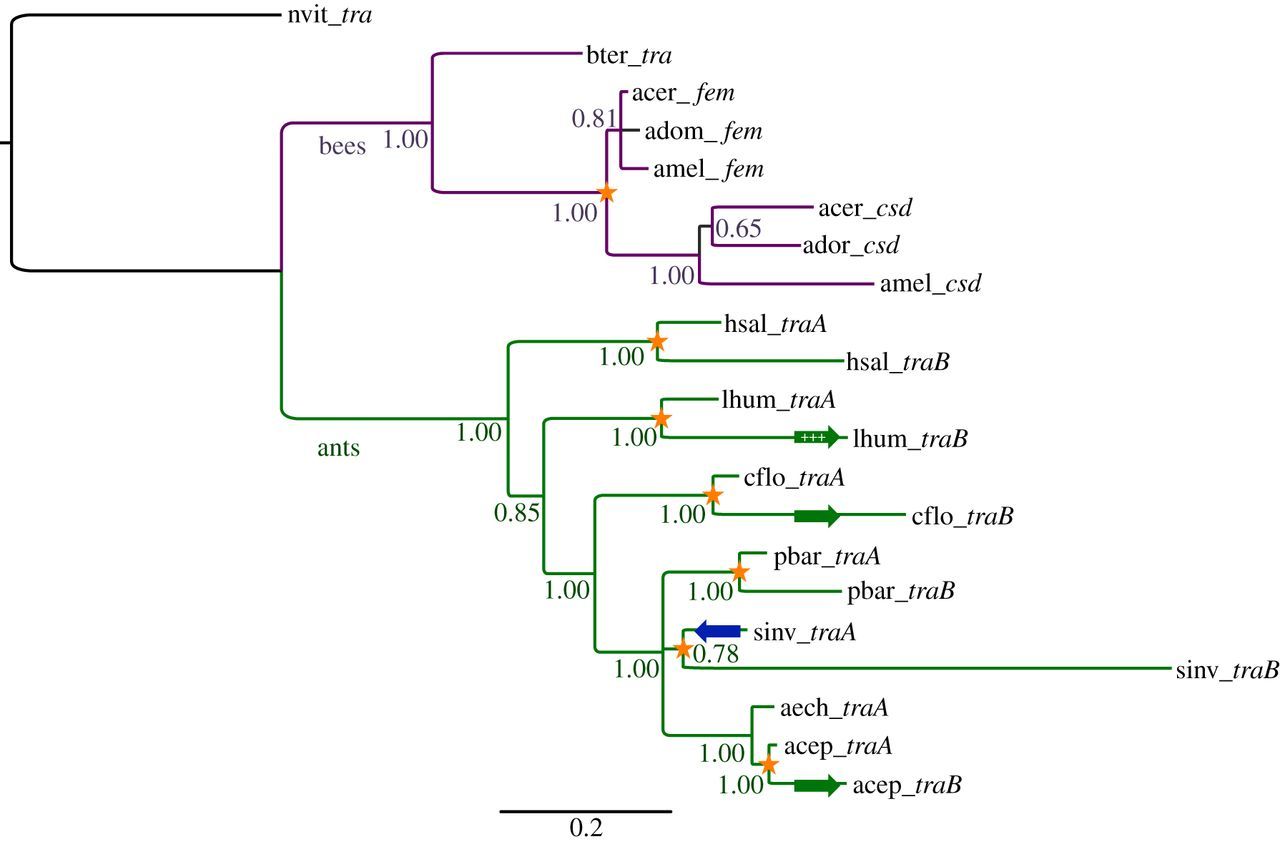
Comparative sequence analyses provided evidence for recombination between tra paralogues, in support of the hypothesis of concerted evolution. Phylogenetic network reconstruction revealed many alternative tree topologies, consistent with the presence of recombinant fragments in the paralogues (figure 2a). A more focused analysis of the leafcutter ant clade that contains the most closely related ant sequences of At. cephalotes and Ac. echinatior (and therefore those with the clearest phylogenetic signal) showed high bootstrap support for two different gene trees (figure 2b). This indicates a recombination event between the paralogues of At. cephalotes (there is only one homologue in Ac. echinatior). The use of the nine statistical methods for the detection of specific recombination events (implemented in the RDP4 package [28]) also provided strong support for a recombination event (by seven of nine methods) in At. cephalotes in a 497 bp segment of the 3’ region of the gene (table 2 and figure 3a). Importantly, the occurrence of inter-locus recombination is strongly supported by three methods having higher detection power compared with the other methods—RDP, Maxchi and Chimaera [33,45]. We further validated this inference using a likelihood-based phylogenetic test (the AU test). The gene tree topology inferred from the putative recombinant region was incompatible with the non-recombinant region of the alignment (p = 0.035) while the topology from the non-recombinant region was not significantly incompatible with the recombinant region (p = 0.17). A statistically significant signature of specific recombination events could not be detected in the other species either because of a genuine lack of recombination or because the divergence time and evolutionary distances between these pairs of paralogues and the nearest orthologues in other species was too large to allow detection of recombination.
Table 2 – Evidence for recombination between tra paralogues in honeybees and in ants. (Analyses were based on coding sequence alignments. The Ap. mellifera recombination event involves the fem sequence AAS86667 and the csd sequence AY352276.)
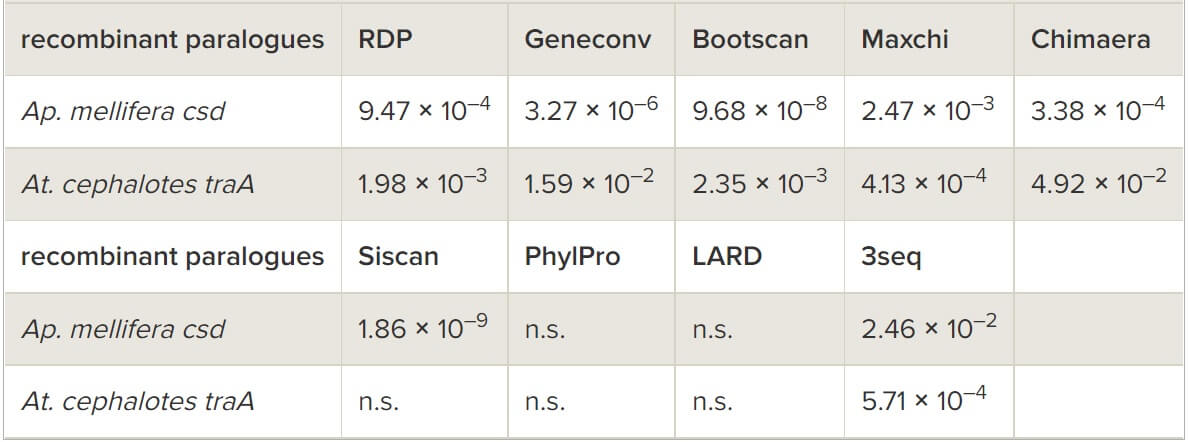
Figure 2
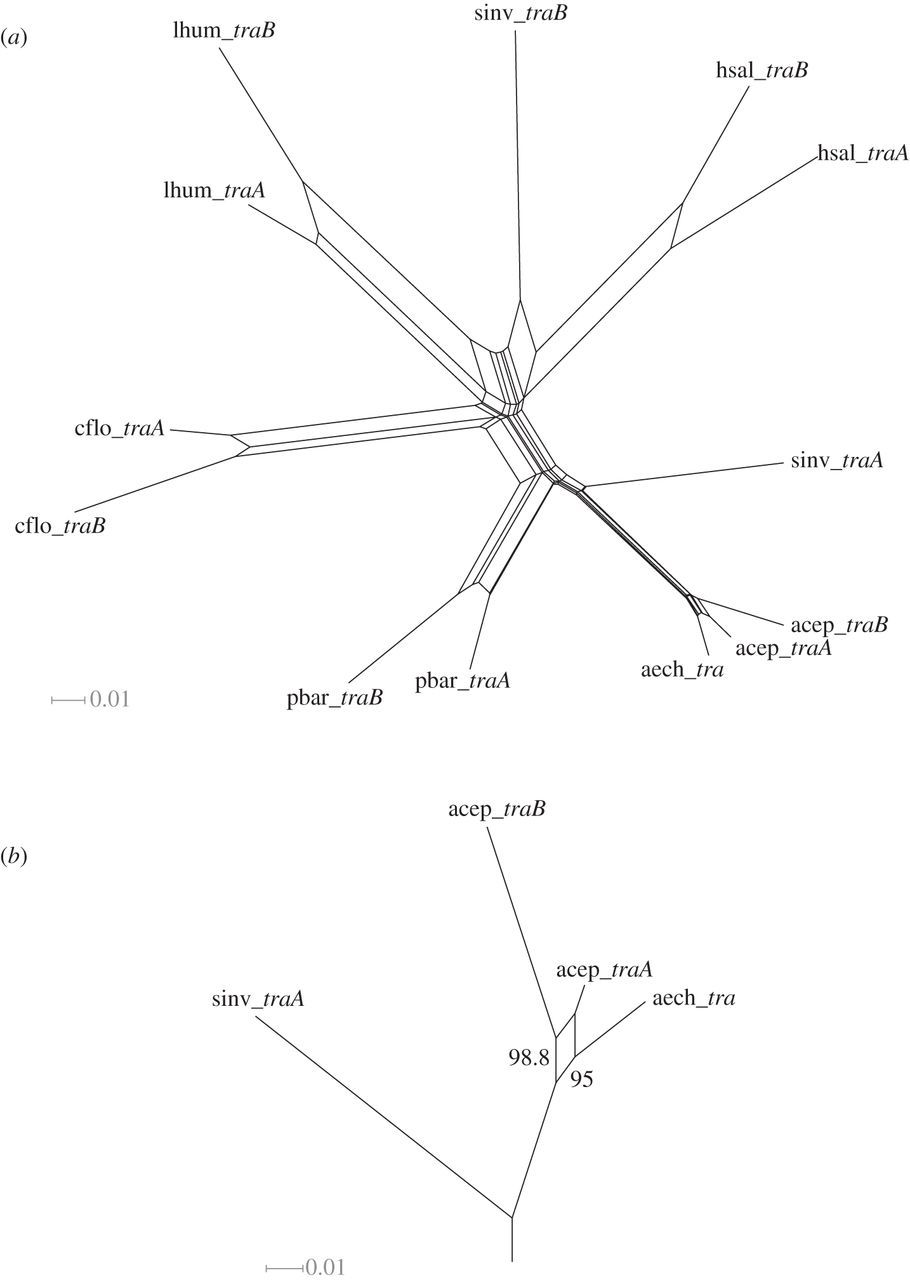
Evidence for recombination between tra paralogues in ants from phylogenetic splits networks constructed using SplitsTree for coding sequences from (a) the seven ant species and for (b) a narrowed dataset of the attine ants (Ac. echinatior and At. cephalotes) with S. invicta traA as an outgroup. Parallelogram branches represent alternative tree topologies corresponding to putative recombination events. Bootstrap support for two alternative topologies is indicated on the parallelogram. Species name abbreviations as in figure 1.
Figure 3
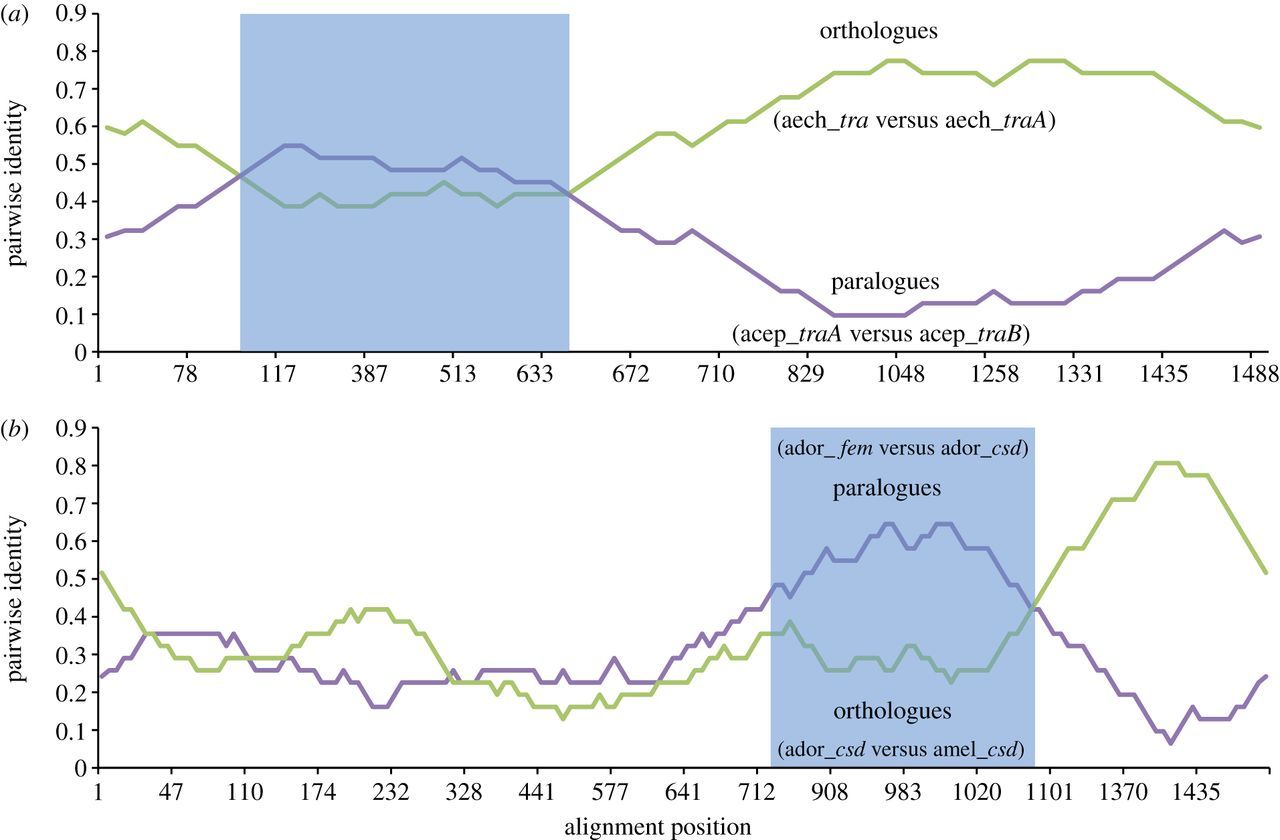
Pairwise identity plots calculated by RDP for the tra coding sequences from (a) the two leaf cutter ant species At. cephalotes and Ac. echinatior and from (b) the two honeybee species Ap. mellifera (csd allele AY352276, fem AAS86667) and Ap. dorsata (csd allele EU100926). Shaded area indicates the putative recombinant region.
We applied the same tests to the paralogue pairs (csd and fem) in the three Apis honeybee species. In contrast to ants, a phylogenetic network reconstructed for the bees paralogues gave low bootstrap support for alternative topology (see the electronic supplementary material, figure S1), implying an ancestral duplication before speciation of the three Apis species. Nevertheless, evidence of recombination and gene conversion was found by several test statistics. We used the suite of nine tests to search for gene conversion events in 79 alleles of the csd loci in the three Apis species. We applied the tests to 100 sequence sets that each included a different random choice of one csd allele for each species. In 87 of these samples, at least one of the test statistics was significant (figure 4a). In particular, RDP, Maxchi, Chimaera and Bootscan found recombination in 39, 80, 67 and 48 samples, respectively. In total, 121 gene conversion events were inferred (77 in Ap. dorsata, 25 in Ap. mellifera and 19 in Ap. cerana; figure 4b, electronic supplementary material, S2 and S3), providing strong evidence that gene conversion occurred between the fem and csd genes in the three honeybee lineages.
Figure 4
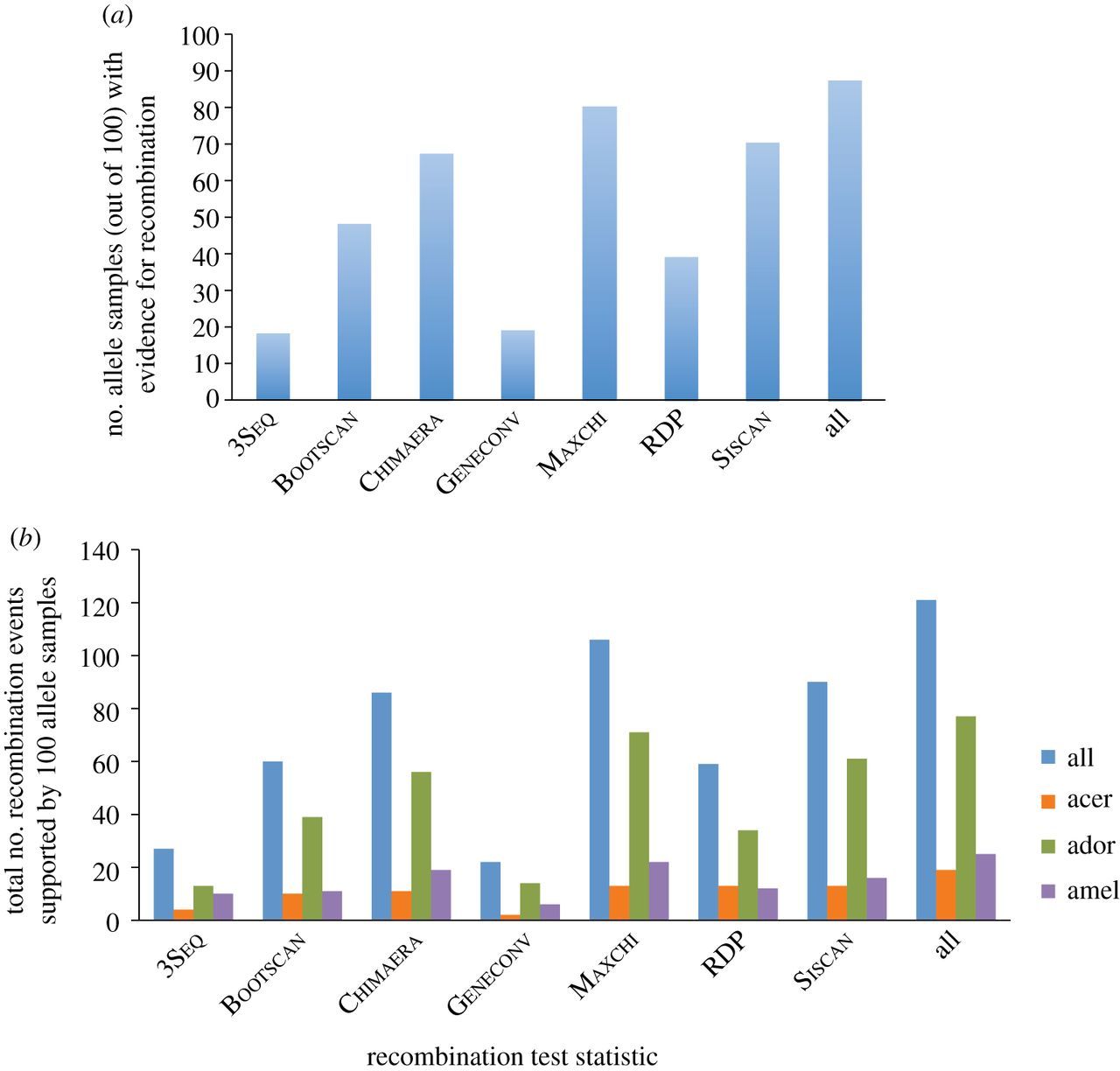
Recombination test statistics applied to fem sequences and samples of csd alleles of Ap. mellifera, dorsata and cerana. (a) The number of samples that received a positive result for each test statistic. (b) The number of instances of inferred recombination events in these samples classified by test and species.
We validated these inferred recombinant csd alleles with the phylogenetic AU test. We applied the AU test to the 48 samples that received a significant score from the Bootscan method, because this indicates a phylogenetic difference between the putative recombinant and the non-recombinant regions of the alignment. These included 63 Bootscan-supported putative gene conversion events. The gene tree topology inferred from the recombinant region was significantly incompatible with the non-recombinant region of the alignment in 17 of the samples while the topology from the non-recombinant region was significantly incompatible with the recombinant region in two of the samples (FDR adjusted p < 0.05; see the electronic supplementary material, table S2). Thus, a robust phylogenetic signal in the recombinant regions of the high-scoring alleles supports the results of the suite of test statistics.
Further support for the inference of gene conversion is found in the form of fem-specific substitutions that were copied to the csd allele. We searched for such substitutions in the top scoring alignment, which contains the putatively recombinant Ap. mellifera csd allele AY352276. A recombination event between Ap. mellifera fem and this csd allele was inferred (by seven of nine methods) in a 207 bp segment at the 3’ end of the gene (table 2). Figure 3b shows the RDP results for this alignment, plotting the pairwise similarity between the Ap. mellifera csd allele, the Ap. mellifera fem and an Ap. cerana csd allele. In the inferred recombinant region the Ap. mellifera csd allele shows a higher similarity to the paralogous Ap. mellifera fem than to the orthologous Ap. cerana csd. We found three substitutions in this region where this Ap. mellifera csd allele shares the same derived nucleotide occurring in the Ap. mellifera fem sequence but not in any of the other csd alleles. No such substitutions are found in the non-recombinant region of the alignment (Fisher’s exact test p = 0.01). This suggests a gene conversion event copying this region from fem to csd in the Ap. mellifera lineage.
In spite of this evidence for gene conversion, the Apis gene tree suggests duplication prior to speciation of the three Apis species (figure 1). This pattern contrasts with the ant gene tree that implies multiple duplications after the ants’ speciation. This difference can be explained by the much greater divergence time between the ants than the bees used in this study. The ant lineages began to speciate more than 115 million years ago [46] while the honeybees speciated less than 40 million years ago [47]. As a result, gene conversion events in honeybees may be limited to a smaller subset of the gene sequence, and thus not yet result in sufficient homogenization among paralogues to make them more similar than orthologues. The same explanation may account for the abovementioned difference in the phylogenetic network results for ants and bees.
(b) Adaptive evolution of tra paralogues
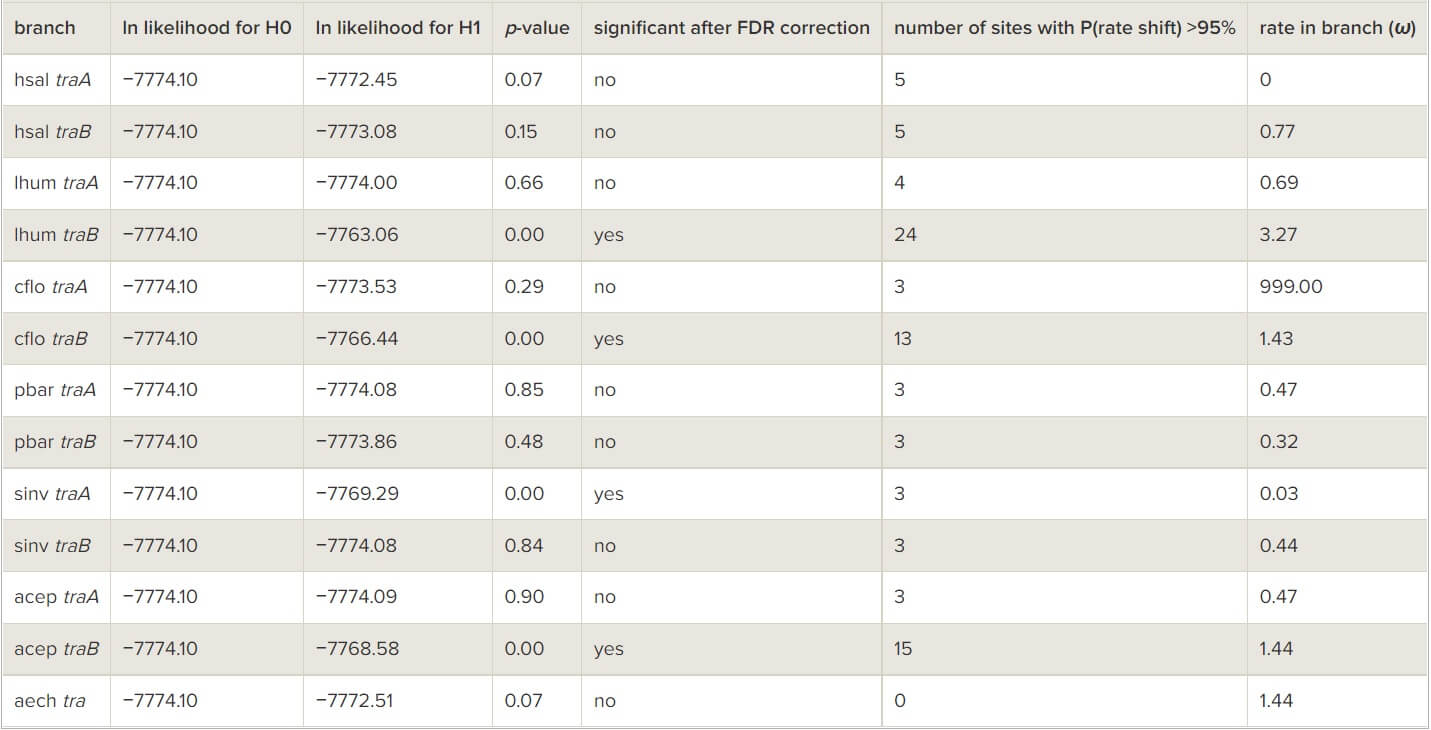
Two lines of evidence indicate that natural selection acts in a similar manner on tra homologues in ants as in bees. First, tra paralogues are adjacent loci in the genomes of the three Apis species [7], as well as in four out of five ant species that have two paralogues with known relative position (in the fifth species the paralogues are separated by seven genes; in the sixth they were found on different scaffolds in the genome assembly so their relative position is unknown; table 1). In ants, the close location is unlikely to be owing to chance alone because extensive genomic rearrangements occurred in these lineages: a comparative analysis of the genomes of the four species where the two tra paralogues are adjacent reveals that only 1.07 per cent of neighbouring pairs of genes in any of these four species remain adjacent in all four genomes (see the electronic supplementary material, table S1). This indicates selection for maintaining synteny of the paralogues. Such physical proximity between tra paralogues probably facilitates concerted evolution [48]. Second, there is evidence for adaptive sequence evolution in tra sequences in ants as was previously described in bees [8]. Branch-site tests provided evidence for positive selection (dN/dS > 1) in at least three codon sites in Linepithema humile traB (table 3) and accelerated evolutionary rate in at least 24, 13 and 15 codon sites in the traB sequences of L. humile, C. floridanus and At. cephalotes, respectively (table 4). Thus, selection appears to be a significant driver of sequence divergence between the two paralogues in both ants and bees.
Table 3 – Tests for positive selection on tra homologues in ants using the PAML branch-site model A. (Species abbreviations are identical to those used in figure 1. Statistically significant results are indicated in bold.)
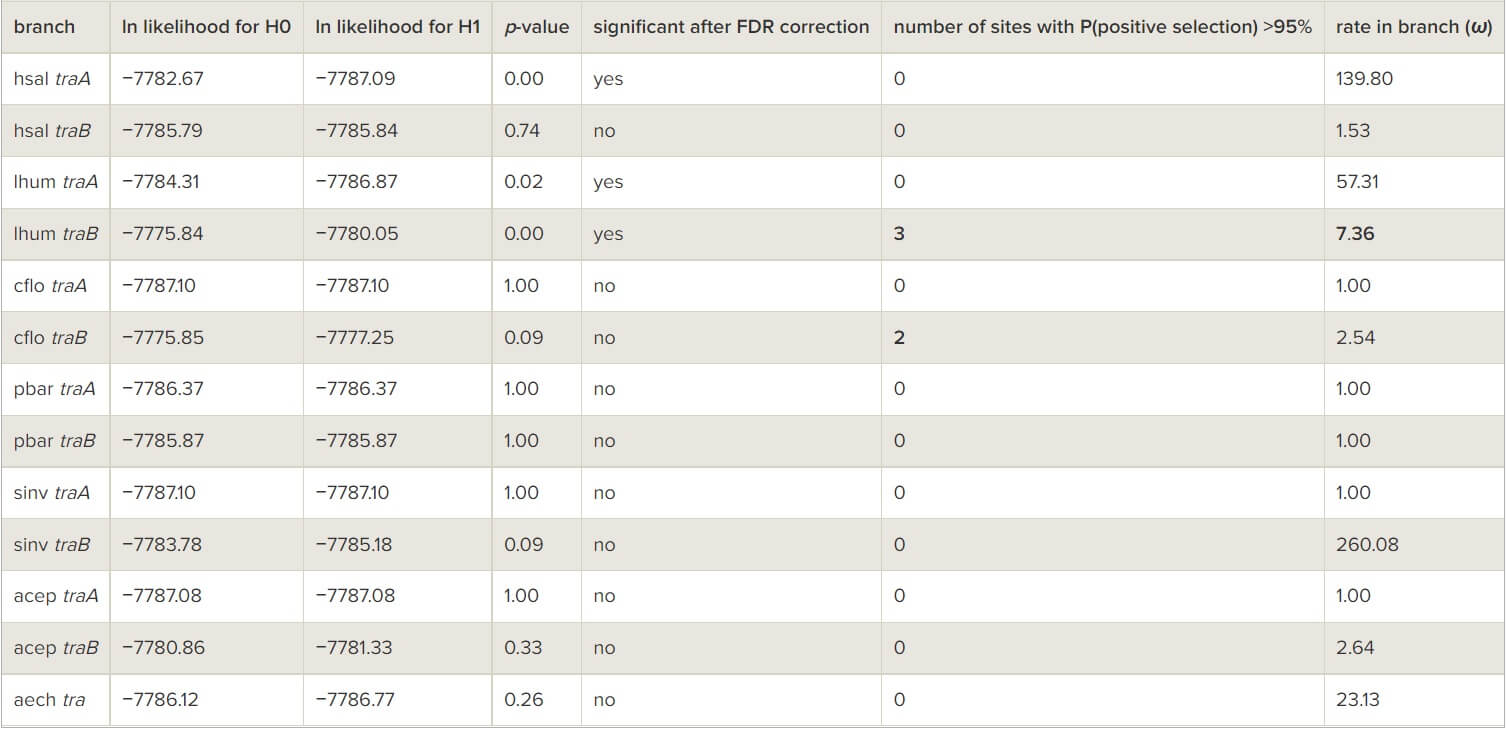
Table 4 – Tests for shifts in the evolutionary rate of tra homologues in ants using the PAML clade model C. (Species abbreviations are identical to those used in figure 1.)
4. Discussion
The comparison of ant genomes revealed the presence of two tra copies in six of the seven species, with the two paralogues being invariably more similar within- than between-species. This striking pattern is inferred to result from the action of concerted evolution over at least 115 million years of evolution. While the alternative hypothesis of multiple independent duplications cannot be completely ruled out, three lines of evidence support the view that gene conversion between the two paralogues has occurred in both ants and bees. First, gene conversion events were found in the ant and bee sequences with support from multiple different recombination test statistics. Second, likelihood-based phylogenetic tests confirmed that recombinant sequences support a different tree topology than the non-recombinant sequences. Third, we identified fem-specific substitutions that are also found in a csd allele of Ap. mellifera. In the light of these results, we interpret the ant gene tree as an ancestral duplication at least 115 million years ago followed by concerted evolution between the two paralogues. The detection of relatively recent gene conversion events suggests that concerted evolution is acting over long evolutionary time in this gene family.
Two lines of evidence suggest that selection pressures are responsible for the evolutionary patterns that we observe. First, the longstanding action of concerted evolution and the conservation of the paralogues in near proximity to one another suggest selection to maintain the ability of paralogues to recombine. Second, we show that one of two paralogues experienced accelerated evolution and/or positive selection in several ant and bee lineages. Hence, we hypothesize that gene conversion between paralogues serves as a mechanism to generate novel recombinant alleles of the fast-evolving paralogue. Novel alleles would be selected for under balancing selection, which is known to act on the csd loci of honeybees.
Selection for concerted evolution at a locus under balancing selection may seem counterintuitive because concerted evolution is better known for homogenizing gene sequences, as in ribosomal RNA genes [44]. However, a diversifying effect of recombination has been previously described in antigens of pathogenic bacteria and protozoa (e.g. [49,50], reviewed in [51]), in vertebrate major histocompatibility complex immune genes [52–54], and in metazoan prdm9 genes [55], all of which are under balancing selection [55–58]. Inter-allelic recombination and gene conversion was first proposed to contribute to the extremely high polymorphism of vertebrate MHC genes (reviewed in [53,54]) and was subsequently shown to generate novel alleles more rapidly than point mutations in birds [52].
Schmieder et al. [59] recently conducted similar analyses to those reported here. An important difference in our studies is that we used multiple methodologies, which are more conservative and robust to artefacts. The use of more conservative methodologies is important because the highly variable and complex patterns of sequence evolution in this gene family could conceivably lead to false inference of recombination. tra paralogues in different lineages were found to evolve with considerably different evolutionary rates, including episodes of positive selection. At least in the three Apis species where multiple allele sequences are available, one of the paralogues (csd) was shown to evolve under strong balancing selection, which generates a pool of highly diverged alleles. Moreover, considerable portions of the protein are repetitive sequences, which are notoriously difficult to align, which may lead to artefacts in molecular evolutionary analyses such as positive selection inference [40,60].
Because of the problems mentioned above, we took special care in our choice of methodologies, aiming for robustness to potential artefacts. There were three important differences between our analyses and those done by Schmieder et al. [59]. First, we used a superior Bayesian method for joint alignment and phylogeny reconstruction (Bali-Phy) to minimize artefacts owing to alignment errors. We also masked unreliable parts of the alignments to prevent alignment errors from affecting the inference of recombination and positive selection [40]. This difference may be responsible to the lesser extent of positive selection that we find. Only one ant gene (L. humile traB) had a significant signal in our analyses, compared with five in the results of Schmieder and colleagues. In two other lineages (traB of C. floridanus and At. cephalotes), we found acceleration of the evolutionary rate, but not necessarily positive selection.
Second, while Schmieder and colleagues used Geneconv to test for recombination, we use eight additional test statistics, including RDP, Maxchi and Chimaera that have been shown to have superior detection power compared with other methods [33,45]. Furthermore, our analyses include phylogenetic methods (RDP and Bootscan), which detect different phylogenetic relationships among the sequences in the inferred recombinant and non-recombinant regions of the alignment. Such methods provide important robustness to the variation in selection pressure that we observe among the paralogues. RDP and other phylogenetic methods were shown to be more robust than non-phylogenetic methods in simulation studies involving variation of selection, including positive selection, as well as asymmetric tree topology and sequence divergence [61]. We did not find the gene conversion events reported by Schmieder and colleagues in the longer branches of the ant phylogeny (Harpegnathos saltator, C. floridanus and P. barbatus), perhaps as a result of the use of these more conservative methodologies. However, our analyses allow one to confidently infer gene conversion in the shorter branches (At. cephalotes).
Third, we validated the inferred recombination events by a superior, likelihood-based phylogenetic test for the significance of the topological difference between the recombinant and the non-recombinant regions. These methodologies are more robust to potential artefacts, thus providing greater confidence in the inference of concerted evolution.
Other differences relative to the study by Schmieder and colleagues include our testing of the entire pool of Apis csd alleles, which allowed robust detection of multiple gene conversion events in all three Apis species, and the identification of an Ap. mellifera csd allele with a clear pattern of gene conversion supported by unique derived substitutions. The inclusion of the second S. invicta paralogue and the two L. humile paralogues provided further evidence for positive selection and better resolution of the phylogeny.
Gene conversion between the paralogues could have interesting implications in terms of their molecular function. Molecular studies in honeybees demonstrated that zygosity of csd regulates alternative splicing of fem mRNA [8], and it has been proposed that the mechanism by which heterozygosity of the csd gene affects fem involves the formation of a heterodimer of csd protein products [62]. Gene conversion events such as the one reported here in the Ap. mellifera csd allele may alter the specificity of the variable C-terminal domains of the protein (RS repeat and proline-rich domains), which were suggested to participate in dimer formation [62]. Thereby, partial gene conversion events could generate novel alleles with altered binding specificities.
Other partial gene conversion events, as exemplified by the At. cephalotes paralogues, may affect the N-terminal domain of the protein. In contrast to the variability of the C-terminal domains within- and between-species, the N-terminal domain is highly conserved among all ant and bee homologues. Such a contrasting pattern is comparable to the contrast between purifying selection at the fem locus and positive selection at the neighbouring csd locus. Owing to their tight linkage, positive selection on csd could lead to a selective sweep (‘hitchhiking effect’) of linked deleterious alleles that may arise in fem. The hitchhiking effect associated with positive selection on csd was even observed in other loci farther away from fem and csd [63]. Therefore, selection is expected to favour recombinant haplotypes, where the linkage between the positively and negatively selected alleles is broken. Similarly, positive selection on the C-terminal domains of csd and traB could lead to a sweep of linked deleterious mutations in the N-terminal domain. Therefore, gene conversion events that copy the conserved sequence of the N-terminal domain from the slow-evolving paralogue (fem/traA) onto the fast-evolving paralogue (csd/traB) may act as a mechanism rescuing the deteriorating molecular function of this domain.
In conclusion, this study provides strong support for concerted evolution based on comparative sequence analyses of the two tra paralogues in ants and bees. We also find evidence for natural diversifying selection acting on one gene in each pair of paralogues, possibly resulting from balancing selection. These observations suggest that concerted evolution may be an important mechanism for diversifying the allele pool of genes under balancing selection.
Acknowledgements
This work was supported by several grants from the Swiss NSF and an ERC advanced grant. We thank Darren Martin for advice on the use of RDP4 and on the robustness of recombination inference to variation of selection. We thank Martin Beye, Leo Beukeboom and two reviewers for their useful comments on the manuscript. We thank Sanne Nygaard for input at early stages of this work.
References
-
Hoshijima K, Inoue K, Higuchi I, Sakamoto H& Shimura Y. 1991 Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252, 833–836.doi:10.1126/science.1902987 (doi:10.1126/science.1902987).
-
Gempe T& Beye M. 2011 Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 33, 52–60.doi:10.1002/bies.201000043 (doi:10.1002/bies.201000043).
-
van Wilgenburg E, Driessen G& Beukeboom LW. 2006 Single locus complementary sex determination in Hymenoptera: an ‘unintelligent’ design? Front Zool. 3, 1.doi:10.1186/1742-9994-3-1 (doi:10.1186/1742-9994-3-1).
-
Whiting PW. 1933 Selective fertilization and sex determination in Hymenoptera. Science 78, 537–538.doi:10.1126/science.78.2032.537-a (doi:10.1126/science.78.2032.537-a).
-
Cook JM. 1993 Sex determination in the Hymenoptera: a review of models and evidence. Heredity 71, 421–435.doi:10.1038/hdy.1993.157 (doi:10.1038/hdy.1993.157).
-
Hasselmann M& Beye M. 2004 Signatures of selection among sex-determining alleles of the honey bee. Proc. Natl Acad. Sci. USA 101, 4888–4893.doi:10.1073/pnas.0307147101 (doi:10.1073/pnas.0307147101).
-
Beye M, Hasselmann M, Fondrk MK, Page RE& Omholt SW. 2003 The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114, 419–429.doi:10.1016/S0092-8674(03)00606-8 (doi:10.1016/S0092-8674(03)00606-8).
-
Hasselmann M, Gempe T, Schiott M, Nunes-Silva CG, Otte M& Beye M. 2008 Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454, 519–522.doi:10.1038/nature07052 (doi:10.1038/nature07052).
-
Bonasio R, et al. 2010 Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071.doi:10.1126/science.1192428 (doi:10.1126/science.1192428).
-
Nygaard S, et al. 2011 The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 21, 1339–1348.doi:10.1101/gr.121392.111 (doi:10.1101/gr.121392.111).
-
Suen G, et al. 2011 The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 7, e1002007.doi:10.1371/journal.pgen.1002007 (doi:10.1371/journal.pgen.1002007).
-
Wurm Y, et al. 2011 The genome of the fire ant Solenopsis invicta. Proc. Natl Acad. Sci. USA 108, 5679–5684.doi:10.1073/pnas.1009690108 (doi:10.1073/pnas.1009690108).
-
Smith CR, et al. 2011 Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl Acad. Sci. USA 108, 5667–5672.doi:10.1073/pnas.1007901108 (doi:10.1073/pnas.1007901108).
-
Smith CD, et al. 2011 Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl Acad. Sci. USA 108, 5673–5678.doi:10.1073/pnas.1008617108 (doi:10.1073/pnas.1008617108).
-
Belote JM& Baker BS. 1982 Sex determination in Drosophila melanogaster: analysis of transformer-2, a sex-transforming locus. Proc. Natl Acad. Sci. USA 79, 1568–1572.doi:10.1073/pnas.79.5.1568 (doi:10.1073/pnas.79.5.1568).
-
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W& Lipman DJ. 1997 Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.doi:10.1093/nar/25.17.3389 (doi:10.1093/nar/25.17.3389).
-
Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sanchez Alvarado A& Yandell M. 2008 Maker: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196.doi:10.1101/gr.6743907 (doi:10.1101/gr.6743907).
-
Lewis SE, et al. 2002 Apollo: a sequence annotation editor. Genome Biol. 3, RESEARCH0082.
-
Redelings BD& Suchard MA. 2005 Joint Bayesian estimation of alignment and phylogeny. Syst. Biol. 54, 401–418.doi:10.1080/10635150590947041 (doi:10.1080/10635150590947041).
-
Zhang J, Nielsen R& Yang Z. 2005 Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479.doi:10.1093/molbev/msi237 (doi:10.1093/molbev/msi237).
-
Yang Z. 2007 PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591.doi:10.1093/molbev/msm088 (doi:10.1093/molbev/msm088).
-
Benjamini Y& Hochberg Y. 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. Crossref,
-
Bielawski JP& Yang Z. 2004 A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59, 121–132.doi:10.1007/s00239-004-2597-8 (doi:10.1007/s00239-004-2597-8).
-
Yang Z, Nielsen R, Goldman N& Pedersen AM. 2000 Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449.
-
Bryant D& Moulton V. 2004 Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265.doi:10.1093/molbev/msh018 (doi:10.1093/molbev/msh018).
-
Huson DH& Bryant D. 2006 Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267.doi:10.1093/molbev/msj030 (doi:10.1093/molbev/msj030).
-
Dress AW& Huson DH. 2004 Constructing splits graphs. IEEE/ACM Trans. Comput. Biol. Bioinform. 1, 109–115.doi:10.1109/TCBB.2004.27 (doi:10.1109/TCBB.2004.27).
-
Martin DP, Lemey P, Lott M, Moulton V, Posada D& Lefeuvre P. 2010 RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–2463.doi:10.1093/bioinformatics/btq467 (doi:10.1093/bioinformatics/btq467).
-
Martin D& Rybicki E. 2000 RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563.doi:10.1093/bioinformatics/16.6.562 (doi:10.1093/bioinformatics/16.6.562).
-
Smith JM. 1992 Analyzing the mosaic structure of genes. J. Mol. Evol. 34, 126–129.doi:10.1007/BF00182389 (doi:10.1007/BF00182389).
-
Padidam M, Sawyer S& Fauquet CM. 1999 Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225.doi:10.1006/viro.1999.0056 (doi:10.1006/viro.1999.0056).
-
Martin DP, Posada D, Crandall KA& Williamson C. 2005 A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 21, 98–102.doi:10.1089/aid.2005.21.98 (doi:10.1089/aid.2005.21.98).
-
Posada D& Crandall KA. 2001 Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl Acad. Sci. USA 98, 13 757–13 762.doi:10.1073/pnas.241370698 (doi:10.1073/pnas.241370698).
-
Gibbs MJ, Armstrong JS& Gibbs AJ. 2000 Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16, 573–582.doi:10.1093/bioinformatics/16.7.573 (doi:10.1093/bioinformatics/16.7.573).
-
Boni MF, Posada D& Feldman MW. 2007 An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176, 1035–1047.doi:10.1534/genetics.106.068874 (doi:10.1534/genetics.106.068874).
-
Holmes EC, Worobey M& Rambaut A. 1999 Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 16, 405–409.doi:10.1093/oxfordjournals.molbev.a026121 (doi:10.1093/oxfordjournals.molbev.a026121).
-
Weiller GF. 1998 Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol. Biol. Evol. 15, 326–335.doi:10.1093/oxfordjournals.molbev.a025929 (doi:10.1093/oxfordjournals.molbev.a025929).
-
Loytynoja A& Goldman N. 2005 An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA 102, 10 557–10 562.doi:10.1073/pnas.0409137102 (doi:10.1073/pnas.0409137102).
-
Penn O, Privman E, Landan G, Graur D& Pupko T. 2010 An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 27, 1759–1767.doi:10.1093/molbev/msq066 (doi:10.1093/molbev/msq066).
-
Privman E, Penn O& Pupko T. 2012 Improving the performance of positive selection inference by filtering unreliable alignment regions. Mol. Biol. Evol. 29, 1–5.doi:10.1093/molbev/msr177 (doi:10.1093/molbev/msr177).
-
Shimodaira H. 2002 An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508.doi:10.1080/10635150290069913 (doi:10.1080/10635150290069913).
-
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W& Gascuel O. 2010 New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321.doi:10.1093/sysbio/syq010 (doi:10.1093/sysbio/syq010).
-
Shimodaira H& Hasegawa M. 2001 CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247.doi:10.1093/bioinformatics/17.12.1246 (doi:10.1093/bioinformatics/17.12.1246).
-
Liao D. 1999 Concerted evolution: molecular mechanism and biological implications. Am. J. Hum. Genet. 64, 24–30.doi:10.1086/302221 (doi:10.1086/302221).
-
Posada D. 2002 Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19, 708–717.doi:10.1093/oxfordjournals.molbev.a004129 (doi:10.1093/oxfordjournals.molbev.a004129).
-
Brady SG, Schultz TR, Fisher BL& Ward PS. 2006 Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc. Natl Acad. Sci. USA 103, 18 172–18 177.doi:10.1073/pnas.0605858103 (doi:10.1073/pnas.0605858103).
-
Engel MS. 1998 Fossil honey bees and evolution in the genus Apis (Hymenoptera: Apidae). Apidologie 29, 265–281.doi:10.1051/apido:19980306 (doi:10.1051/apido:19980306).
-
Hsu CH, Zhang Y, Hardison RC, Green ED& Miller W. 2010 An effective method for detecting gene conversion events in whole genomes. J. Comput. Biol. 17, 1281–1297.doi:10.1089/cmb.2010.0103 (doi:10.1089/cmb.2010.0103).
-
Hagblom P, Segal E, Billyard E& So M. 1985 Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315, 156–158.doi:10.1038/315156a0 (doi:10.1038/315156a0).
-
Nystedt B, Frank AC, Thollesson M& Andersson SG. 2008 Diversifying selection and concerted evolution of a type IV secretion system in Bartonella. Mol. Biol. Evol. 25, 287–300.doi:10.1093/molbev/msm252 (doi:10.1093/molbev/msm252).
-
Deitsch KW, Moxon ER& Wellems TE. 1997 Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol. Mol. Biol. Rev. 61, 281–293.
-
Spurgin LG, Van Oosterhout C, Illera JC, Bridgett S, Gharbi K, Emerson BC& Richardson DS. 2011 Gene conversion rapidly generates major histocompatibility complex diversity in recently founded bird populations. Mol. Ecol. 20, 5213–5225.doi:10.1111/j.1365-294X.2011.05367.x (doi:10.1111/j.1365-294X.2011.05367.x).
-
Yeager M& Hughes AL. 1999 Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunol. Rev. 167, 45–58.doi:10.1111/j.1600-065X.1999.tb01381.x (doi:10.1111/j.1600-065X.1999.tb01381.x).
-
Carrington M. 1999 Recombination within the human MHC. Immunol. Rev. 167, 245–256.doi:10.1111/j.1600-065X.1999.tb01397.x (doi:10.1111/j.1600-065X.1999.tb01397.x).
-
Oliver PL, et al. 2009 Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5, e1000753.doi:10.1371/journal.pgen.1000753 (doi:10.1371/journal.pgen.1000753).
-
Aguilar A, Roemer G, Debenham S, Binns M, Garcelon D& Wayne RK. 2004 High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl Acad. Sci. USA 101, 3490–3494.doi:10.1073/pnas.0306582101 (doi:10.1073/pnas.0306582101).
-
Hedrick PW. 1998 Balancing selection and MHC. Genetica 104, 207–214.doi:10.1023/A:1026494212540 (doi:10.1023/A:1026494212540).
-
Smith NH, Maynard Smith J& Spratt BG. 1995 Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol. Biol. Evol. 12, 363–370.
-
Schmieder S, Colinet D& Poirie M. 2012 Tracing back the nascence of a new sex-determination pathway to the ancestor of bees and ants. Nat. Commun. 3, 895.doi:10.1038/ncomms1898 (doi:10.1038/ncomms1898).
-
Fletcher W& Yang Z. 2010 The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol. Biol. Evol. 27, 2257–2267.doi:10.1093/molbev/msq115 (doi:10.1093/molbev/msq115).
-
Bay RA& Bielawski JP. 2011 Recombination detection under evolutionary scenarios relevant to functional divergence. J. Mol. Evol. 73, 273–286.doi:10.1007/s00239-011-9473-0 (doi:10.1007/s00239-011-9473-0).
-
Beye M. 2004 The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. Bioessays 26, 1131–1139.doi:10.1002/bies.20098 (doi:10.1002/bies.20098).
-
Hasselmann M, Lechner S, Schulte C& Beye M. 2010 Origin of a function by tandem gene duplication limits the evolutionary capability of its sister copy. Proc. Natl Acad. Sci. USA 107, 13 378–13 383.doi:10.1073/pnas.1005617107 (doi:10.1073/pnas.1005617107).
Supplemental Material
rspb20122968supp1.pdf (.PDF, 3.2 MB)
Footnotes
© 2013 The Author(s) Published by the Royal Society. All rights reserved.