Ants, Bees, Genomes & Evolution @ Queen Mary University London
Published: 23 October 2014
Convergent Genetic Architecture Underlies Social Organization in Ants
Jessica Purcell, Alan Brelsford, Yannick Wurm, Nicolas Perrin, Michel Chapuisat
Current Biology, 26:2864-79
Highlights
-
Colony queen number in Alpine silver ants is controlled by a large set of linked genes
-
Suppression of recombination maintains this “social chromosome”
-
Ant species converge in the genetic architecture underlying social organization
-
The social chromosomes from two ant species harbor different sets of genes
Summary
Complex adaptive polymorphisms are common in nature, but what mechanisms maintain the underlying favorable allelic combinations [1, 2, 3, 4]? The convergent evolution of polymorphic social organization in two independent ant species provides a great opportunity to investigate how genomes evolved under parallel selection. Here, we demonstrate that a large, nonrecombining “social chromosome” is associated with social organization in the Alpine silver ant, Formica selysi. This social chromosome shares architectural characteristics with that of the fire ant Solenopsis invicta [2], but the two show no detectable similarity in gene content. The discovery of convergence at two levels—the phenotype and the genetic architecture associated with alternative social forms—points at general genetic mechanisms underlying transitions in social organization. More broadly, our findings are consistent with recent theoretical studies suggesting that suppression of recombination plays a key role in facilitating coordinated shifts in coadapted traits [5, 6].
Graphical Abstract
Results and Discussion
The convergent evolution of similar traits in distantly related species demonstrates the power of natural selection [7]. But when complex sets of behavioral and morphological traits arise repeatedly, are they controlled by similar genetic mechanisms? Comparing the genetic architecture underlying convergent adaptations can lead to key insights into how genomic evolution shapes phenotypic innovations.
Many ant species exhibit strikingly convergent behavioral syndromes depending upon the number of queens reproducing in the colony. In general, colonies with multiple queens (polygynous) produce smaller queens and workers than colonies with a single queen (monogynous), and the two forms differ in key behavioral traits such as tolerance of conspecifics and mode of dispersal [8, 9]. A recent study in the fire ant Solenopsis invicta identified a large, nonrecombining “social chromosome” that is associated with alternative social organizations in that species [2]. Here, we investigated the genomic architecture underlying social organization in the Alpine silver ant, Formica selysi, which is polymorphic in queen number and exhibits a similar suite of behavioral and morphological traits associated with each form.
Populations of F. selysi contain a mix of monogynous and polygynous colonies [10, 11], with rare oligogynous colonies headed by two closely related queens (see Table S1 available online; [12]). In our study population in the Swiss Alps, we have monitored the social organization of 121 colonies over the past 14 years [10, 11]. We have identified differences between the two common social forms in body size of workers and queens [12, 13], colony lifespan [13], colony size [13], allocation to reproductive offspring [14], and brood development time [15]. The social forms are not differentiated at eight polymorphic microsatellite loci but instead form a single, apparently genetically homogeneous population [10, 11]. Taken together, these characteristics make this species an ideal system for the investigation of the genetic basis of social organization.
Here, we performed a genome-wide association study to search for genetic markers exhibiting significant allele frequency differences between the two social forms. Through genotyping-by-sequencing [16, 17] of haploid males from monogynous and polygynous colonies within the focal population, we identified 18,199 SNPs (see Experimental Procedures). At a genome-wide false discovery rate of 0.01 (per-SNP α = 0.0003), 643 of these markers were significantly associated with social organization in our mixed-effects model (Table S2).
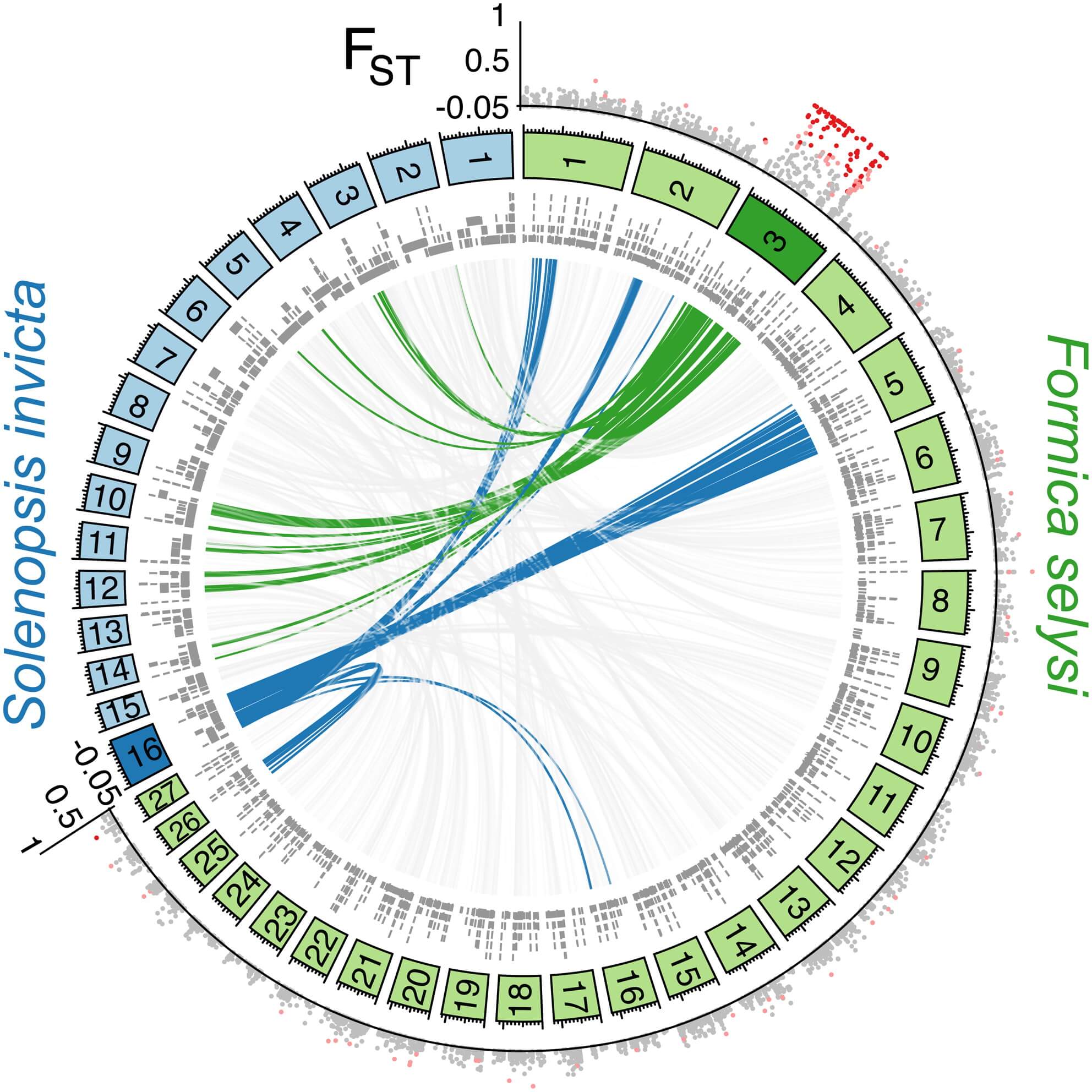
To identify the position of these markers in the genome, we constructed a linkage map (see Experimental Procedures). This map contained 27 main linkage groups, consistent with the haploid chromosome number of members of the Serviformica subgenus (Figure 1A; [18]). A total of 2,409 markers, located on 1,763 genome scaffolds, were heterozygous in the focal queen. These scaffolds contained 36.7% of the SNPs from the population data set. Strikingly, of the 136 SNP markers associated with social organization that could be placed on the linkage map, 134 were located on a single linkage group (Figure 1A). Only two markers did not map to this linkage group, a number consistent with the number of false positives expected given the false discovery rate we used. These two markers each mapped to a different linkage group. The linkage group (LG3) containing highly differentiated SNPs between males of monogynous and polygynous origin is hereafter called the “social chromosome.” The differentiated SNPs occurred across most of the linkage group (183 of 244 cM), with short sections of low differentiation characterizing each end (Figure 1B).
Figure 1 – The Social Chromosomes of F. selysi and S. invicta Are Not Homologous
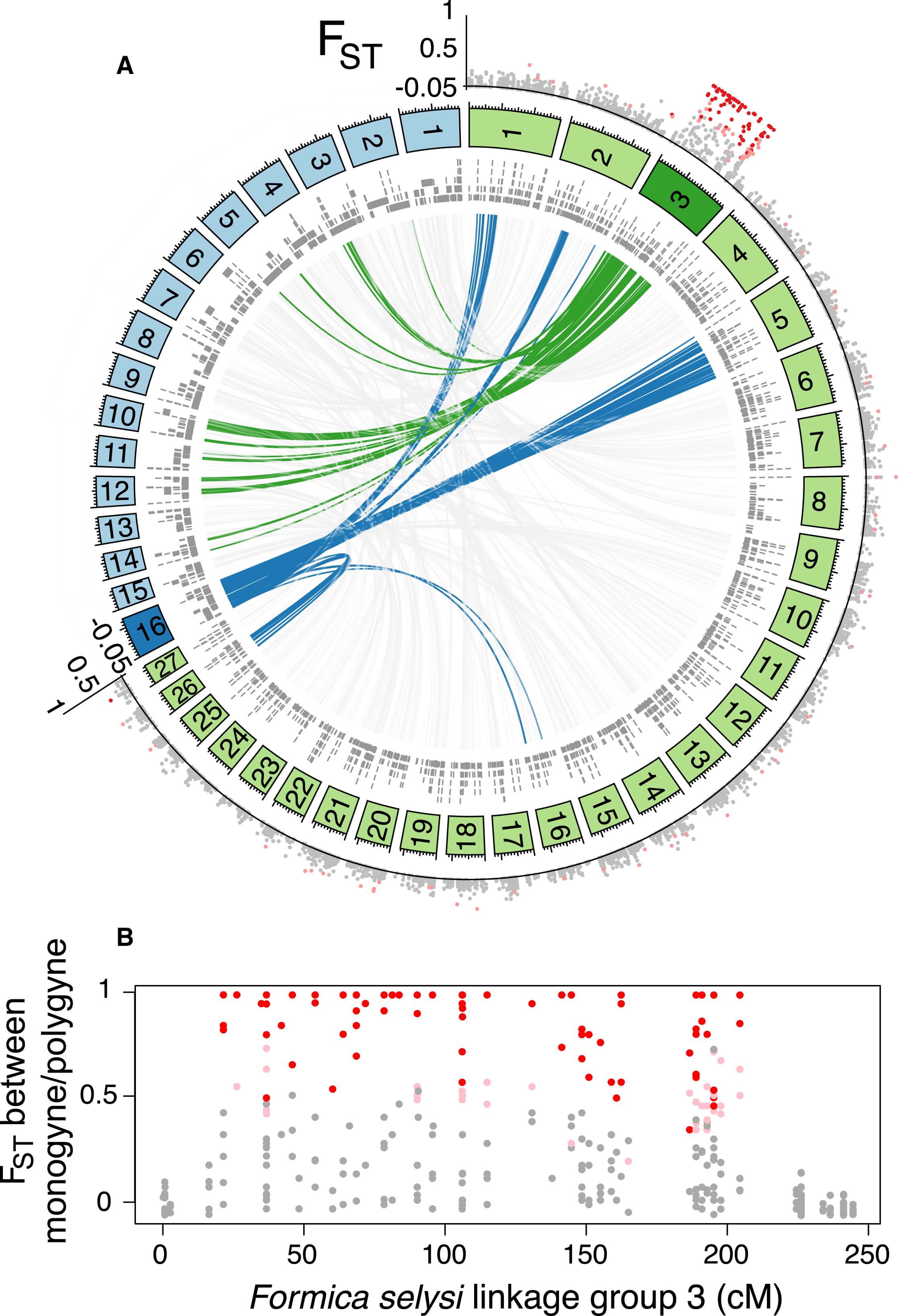
Markers with a high degree of differentiation between monogynous and polygynous males were localized on one F. selysi linkage group not homologous to the fire ant social chromosome. The circle in (A) shows linkage groups of F. selysi (green panels) and S. invicta (blue panels), with scaffolds from respective genomes aligning to loci in each linkage group (gray bars, inner circle). The interior lines show synteny between the two genomes for the F. selysi social chromosome (green), the S. invicta social chromosome (blue), and scaffolds on other linkage groups (light gray). The FST value between monogyne and polygyne males at each SNP placed on the linkage map is shown in the outer circle, with colors indicating no significant differentiation between the two forms (gray dots), significant differentiation at α = 0.01 (light red dots), and significant differentiation at α = 0.0003 (dark red dots). In (B), a zoomed view of the FST value between monogyne and polygyne males on the F. selysi social chromosome shows the lack of divergence at each end of the chromosome.
We identified two major haplotypes at the social chromosome: the sequence found in males of monogynous origin is designated as the “Sm” haplotype, and the sequence associated with males of polygynous origin is called the “Sp” haplotype. How are these allelic differences maintained over such a large region of the genome? We expected that this pattern could result from suppression of recombination between the two haplotypes, and we investigated this possibility by constructing a linkage map of markers on the social chromosome from offspring of four Sm/Sp heterozygous queens. As predicted, the linkage map showed perfect cosegregation of the Sm versus Sp variants of markers from 36.6 to 195.1 cM along the Sm/Sm linkage map (Figure 2A; see Experimental Procedures). This pattern shows that recombination is suppressed between the two haplotypes in these four families. This genomic region is characterized by high linkage disequilibrium and differentiation between Sm and Sp haplotypes, suggesting a long history of suppressed recombination, beyond the single generation demonstrated with the linkage map (Figures 1, 2, and S1).
Figure 2 – Recombination Is Suppressed over the Central Region of the Social Chromosome
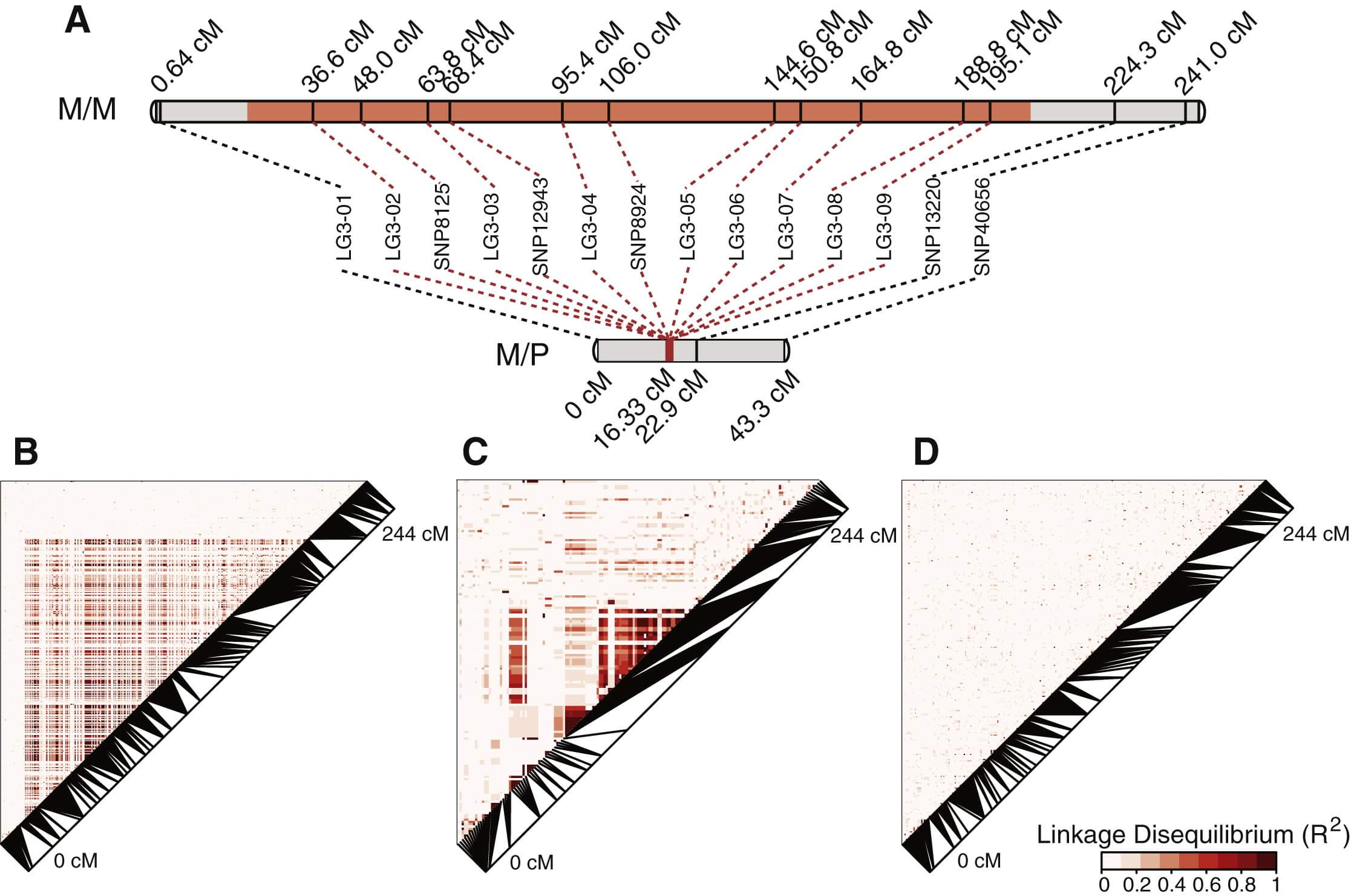
(A) Linkage maps of the social chromosome show suppressed recombination in 80 worker offspring of four Sm/Sp queens relative to 59 male offspring of one Sm/Sm queen; no recombination was observed in offspring of Sm/Sp queens (i.e., map length 0 cM) for markers in the region that is divergent between the Sm and Sp chromosomes, which spans 183 cM in the Sm/Sm linkage map. The region with significantly different Sm and Sp haplotypes is shown in orange, while regions with little differentiation between monogynous and polygynous individuals are shown in gray.
(B–D) Linkage disequilibrium (LD; R2) between markers along linkage group 3 (the social chromosome) is shown between Sm and Sp (B; n = 79 males), within Sp (C; n = 30 males), and within Sm (D; n = 49 males). Between social forms, the area of suppressed recombination shows a higher degree of LD compared to the recombining edges of the linkage group (B). Within Sp (C), the high degree of LD between nonadjacent markers (on the monogyne linkage map) suggests some degree of chromosomal rearrangement and suppression of recombination. No evidence of suppressed recombination is visible in Sm (D).
Identifying genomic regions associated with colony social organization raises immediate questions about the distribution of the two social haplotypes in diploid individuals within and across field colonies. Assessing the genotypic structure of 42 monogynous, 44 polygynous, and 5 oligogynous colonies, we found that monogynous and oligogynous colonies contained only Sm/Sm queens and workers, and Sm males (Table 1). Polygynous colonies contained a combination of Sm/Sp heterozygous and Sp/Sp homozygous queens and workers, as well as Sp males. We never found Sm/Sm queens or workers, or Sm males, in polygynous colonies. Individuals with the Sm/Sm genotype were significantly larger than individuals with a Sp haplotype; there was no difference in size between Sm/Sp and Sp/Sp workers or queens (Table S1; see also [12, 19]).
Table 1. – Genotype and Allele Frequencies of Individuals from Monogynous, Oligogynous, and Polygynous Field Colonies
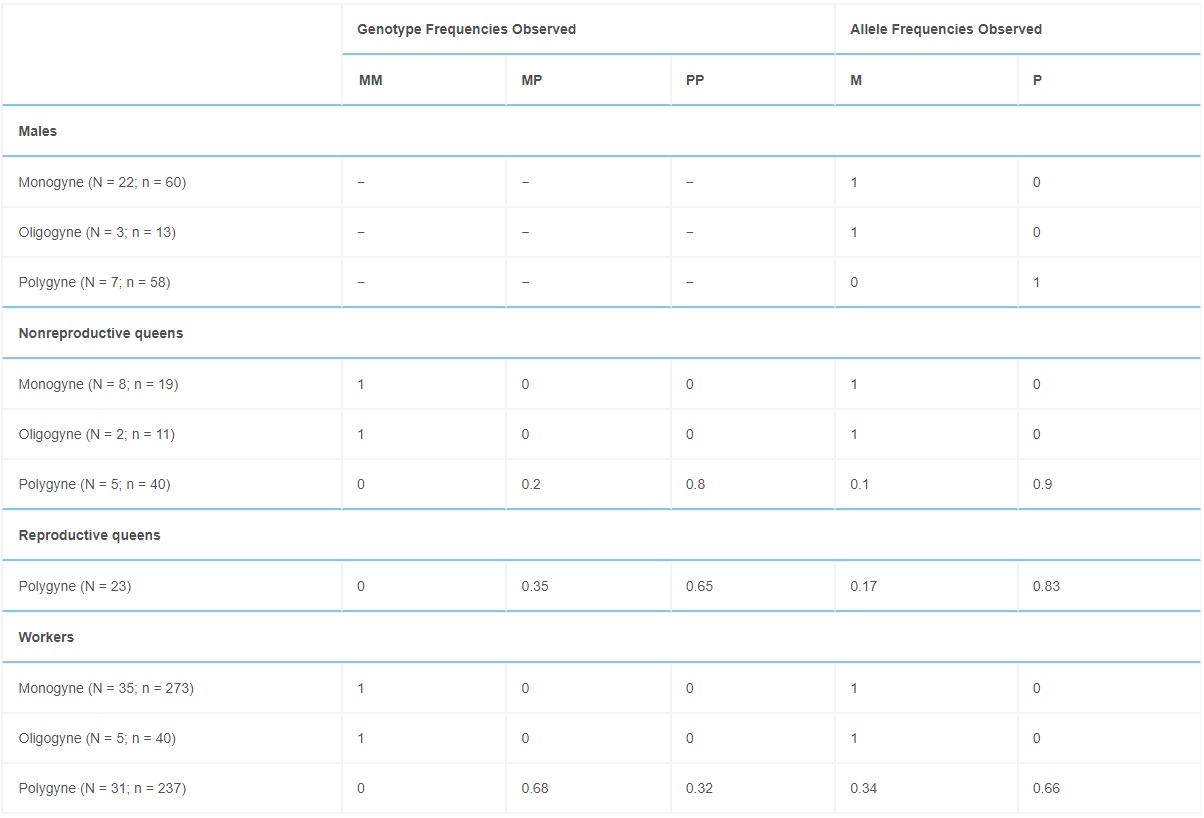
N refers to the number of colonies used; n refers to the total sample of individuals.
The absence of Sm/Sm females and Sm males from polygynous colonies is surprising, since the Sm/Sp queens are expected to produce Sm and Sp male offspring and, when mated with Sm males, heterozygous and Sm/Sm females. The causes of the absence of Sm or Sm/Sm individuals in polygynous colonies deserve further investigation. Possible mechanisms that might generate this unusual genotype distribution include assortative mating (e.g., [20]), meiotic drive (e.g., [21]), differential mortality (e.g., [22]), or brood elimination by workers [23]. More generally, studies on mate choice, brood development, and ecological success will be needed to explain the maintenance of the genetic polymorphism at the social chromosome in this system.
The genotypic system underlying social organization in F. selysi colonies resembles that of fire ants (Table 2). Fire ants exhibit two haplotypes associated with colony queen number: SB and Sb. In both species, monogynous colonies have similar homozygous genotypic compositions at the social chromosome: F. selysi colonies contain only Sm/Sm females mated with Sm males (Table 1), and S. invicta colonies contain only SB/SB females mated with SB males [23]. There are, however, at least two major differences between the two systems. First, polygynous S. invicta colonies contain a mix of SB/SB, SB/Sb, and rare Sb/Sb workers, while all reproductive queens are SB/Sb [24]. In contrast, we found a mix of Sp/Sp and Sm/Sp queens and workers, and no Sm/Sm females or Sm males, in polygynous F. selysi colonies. Second, S. invicta Sb/Sb workers are rare because Sb is generally a recessive lethal allele in females [2, 23]. The F. selysi Sp allele is not lethal in homozygous individuals, as Sp/Sp queens and workers were present in many polygynous colonies (Table 1).
Table 2. – Comparison of the Social Organization of F. selysi and S. invicta
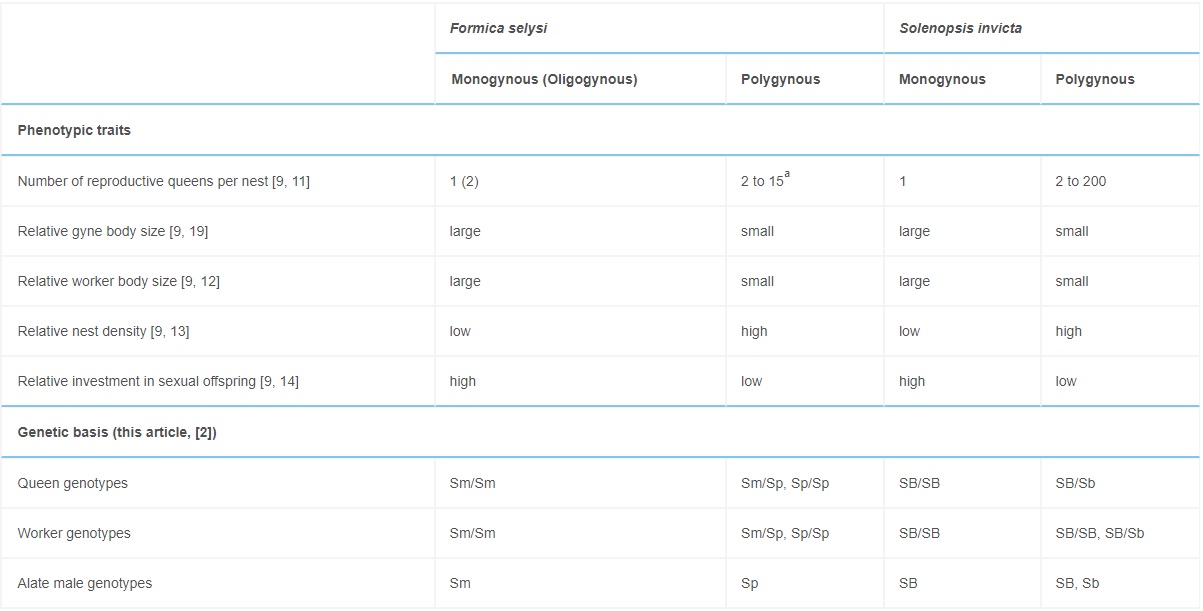
In F. selysi, Sm and Sp refer to the social chromosome haplotypes found in monogynous males and polygynous males, respectively. In S. invicta, the corresponding haplotypes are denoted SB and Sb [2].
a We have observed up to 15 queens in colonies, but it is likely that there are more queens present in highly polygynous colonies, since queens spend most of their time in the subterranean nest chambers, and we only survey the top layer of the nest.
The social chromosome of F. selysi shows structural similarities with that of S. invicta (Figure 1A). In both species, a large nonrecombining region is associated with variation in colony social organization, which includes having one or more queens, as well as multiple correlated morphological and behavioral traits (Table 2). Each nonrecombining region is flanked by genomic regions that recombine between the two haplotypes. Hence, the lineages in which the two species originated, which separated from their common ancestor roughly 130 million years ago [25] and represent distinct origins of polygyny, independently evolved a similar genomic architecture associated with social organization.
Although the two species exhibit similarities in the genetic architecture associated with convergent, multitrait phenotypes, we detected no homology between the social chromosomes of F. selysi and of S. invicta. The scaffolds in the F. selysi social chromosome align to S. invicta scaffolds in seven linkage groups, not including the S. invicta social chromosome, while the F. selysi scaffolds aligning to the S. invicta social chromosome occur in five non-social linkage groups (Figure 1A). Overall, we found no evidence that the social chromosomes of the two species harbor the same set of genes. Despite this general absence of homology, it is still possible that a few genes that were not present on our linkage map are shared between the social chromosomes of F. selysi and S. invicta. Moreover, different transcription factors or other trans-acting elements in each social chromosome may trigger a similar downstream regulatory cascade in the two species. More work is needed to pinpoint the causative mutations and developmental pathways leading to alternative social organization in both species.
The convergence of the genetic architecture underlying F. selysi and S. invicta social organization sheds light on the genomic evolution underlying coordinated shifts in multiple traits. Nonrecombining regions associated with such evolutionary shifts are widely acknowledged to play a central role in speciation and local adaptation (e.g., [26, 27, 28]). The ant social chromosomes show that this genetic architecture is also important for maintaining sympatric polymorphisms. Nonrecombining regions are likely shaped by similar selection pressures, which have been explored in models of coadapted gene complexes, also called supergenes (e.g., [5, 6, 29, 30]). Dobzhansky [29] proposed that zones of suppressed recombination in the genome maintain beneficial combinations of alleles. These diverging allele combinations can be positively selected either in alternative external environments, as in locally adapted populations, or in alternative phenotypes (i.e., when genes harbor alleles with different fitness effects in males and females, or in monogynous and polygynous colonies; e.g., [31]). Suppressed recombination between such genes often involves chromosomal inversions, gene translocations, insertions, or deletions [29] but can also occur in the absence of chromosomal rearrangements (e.g., [32, 33]).
Supergenes underlie very diverse coadapted phenotypes, from the maintenance of two sexes through nonrecombining sex chromosomes [31] to the control of wing coloration and mimicry in Heliconius butterflies [4] and the maintenance of alternative behavioral syndromes in mice and white-throated sparrows [20, 34, 35, 36]. A key difference between the F. selysi social chromosome and many previously studied supergenes indicates that the F. selysi social chromosome follows a distinct evolutionary trajectory. In F. selysi, both social chromosome haplotypes occur in homozygotes. In contrast, in many other systems, one haplotype occurs only in heterozygotes. For example, mice and sparrows strongly prefer to mate with individuals exhibiting the opposite behavioral syndrome [20, 34, 35, 36], and one haplotype tends to be a recessive lethal in both fire ants and mice [2, 35]. Consequently, one haplotype is restricted to heterozygotes, recombination ceases for this haplotype, deleterious mutations accumulate through Muller’s ratchet, and this haplotype tends to degenerate [31] (e.g., the Sb haplotype in fire ants has a higher frequency of repetitive elements and larger introns than the SB haplotype [2]). In F. selysi, viable Sm/Sm and Sp/Sp homozygotes are common, so degeneration of one haplotype is not expected despite the suppression of recombination between Sm and Sp (Figure 2A). We do, however, observe reduced polymorphism in the Sp haplotype compared to the Sm, and further research is required to determine the implications of this pattern (Figure S2). Comparing systems that differ in assortative mating tendencies and haplotype degeneration will provide insights into the factors that initiate chromosomal degeneration in autosomal nonrecombining regions.
Overall, the convergence in genetic architecture and phenotype of two independent socially polymorphic ant species demonstrates that the formation of nonrecombining regions is a key genetic mechanism underlying transitions in social organization in ants. The parallel evolution of nonrecombining regions underlying variation in social systems strongly supports recent theoretical claims that chromosomal rearrangements and the formation of supergenes are important for the evolution of novel phenotypes [5, 6, 37]. Blocks of suppressed recombination would facilitate coordinated shifts in coadapted traits, which may lead to speciation and/or adaptive shifts, or allow for the maintenance of complex phenotypic polymorphisms [3, 4, 38, 39]. The lack of homology between the social chromosomes of two independent ant species further suggests that the genetic systems underlying the convergent evolution of complex phenotypes are not strongly constrained at the level of individual genes. At a larger scale, the similar architecture and independent evolution of social chromosomes and other supergenes point to common principles governing the suppression of recombination in large genomic regions controlling complex coadapted polymorphisms.
Experimental Procedures
A genotyping-by-sequencing approach [16, 17] was used to test the association of markers throughout the genome with social structure and to construct a linkage map for F. selysi. For the association study, the genomes of 79 haploid males from a total of 23 field colonies from a single locality were scanned (2–5 individuals from each of 18 monogynous colonies and 5–6 individuals from each of 5 polygynous colonies). Social structure of all colonies was independently assessed through microsatellite parentage analysis [10, 11]. A linear mixed-effects model was used to test for association between each SNP marker and the social organization of the colony. From 59 male offspring of a single monogynous queen, a linkage map was generated. In complement, a draft genome was produced from a single monogynous male. This genome assembly was used to investigate synteny between F. selysi and S. invicta. Across the social chromosome, nine microsatellite and five SNP markers were developed from the genome, and these were used to prepare a linkage map for 80 worker offspring of four heterozygous queens. Three SNPs that were diagnostic for social organization were further used to assess the genotypic structure of males, queens, and workers from additional F. selysi colonies. Additional males (n = 52), unmated queens (n = 70), reproductive queens (n = 23), and workers (n = 550) from 91 field colonies belonging to the same population were tested with this method.
Detailed methods are provided in the Supplemental Experimental Procedures.
Author Contributions
J.P., A.B., and M.C. designed the study and contributed to all aspects of the project. J.P. and A.B. carried out genetic data collection and bioinformatics analyses. Y.W. provided advice on genetic methods and bioinformatics approaches. N.P. clarified and developed the analogy with models of supergene evolution. J.P., A.B., and M.C. wrote the manuscript with editorial input from all authors.
Acknowledgments
We wish to thank D. Croll, C. Grossen, L. Keller, M. Kirkpatrick, S. Otto, M. Robinson-Rechavi, T. Schwander, S. Yeaman, and four anonymous reviewers for their input on the manuscript and G. Metthez for assistance in the lab. All bioinformatics analyses were carried out on the Vital-IT computer cluster of the Swiss Institute for Bioinformatics. This research was supported by the Fondation Herbette and Swiss National Science Foundation grants 31003A-125306 and 31003A-146641 to M.C. and 31003A-129894 to N.P.
Accession Numbers
Genetic sequences reported herein have been deposited at the NCBI Sequence Read Archive with the accession numbers PRJNA260443 (genome), PRJNA260459 (association study), and PRJNA260462 (linkage map).
Supplemental Information
Download .pdf (1.68 MB)
Document S1. Figures S1 and S2, Tables S1 and S3, and Supplemental Experimental Procedures
Download .xlsx (.66 MB)
Table S2. Related to Figure 1
References
-
Schwander T. Libbrecht R. Keller L. Supergenes and complex phenotypes. Curr. Biol. 2014; 24: R288-R294
-
Wang J. Wurm Y. Nipitwattanaphon M. Riba-Grognuz O. Huang Y.-C. Shoemaker D. Keller L. A Y-like social chromosome causes alternative colony organization in fire ants. Nature. 2013; 493: 664-668
-
Hermann K. Klahre U. Moser M. Sheehan H. Mandel T. Kuhlemeier C. Tight genetic linkage of prezygotic barrier loci creates a multifunctional speciation island in Petunia. Curr. Biol. 2013; 23: 873-877
-
Joron M. Frezal L. Jones R.T. Chamberlain N.L. Lee S.F. Haag C.R. Whibley A. Becuwe M. Baxter S.W. Ferguson L. et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature. 2011; 477: 203-206
-
Yeaman S. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc. Natl. Acad. Sci. USA. 2013; 110: E1743-E1751
-
Kirkpatrick M. Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006; 173: 419-434
-
Wake D.B. Wake M.H. Specht C.D. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science. 2011; 331: 1032-1035
-
Keller L. Social life: the paradox of multiple-queen colonies. Trends Ecol. Evol. 1995; 10: 355-360
-
Ross K.G. Keller L. Ecology and evolution of social organization: insights from fire ants and other highly eusocial insects. Annu. Rev. Ecol. Syst. 1995; 26: 631-656
-
Chapuisat M. Bocherens S. Rosset H. Variable queen number in ant colonies: no impact on queen turnover, inbreeding, and population genetic differentiation in the ant Formica selysi. Evolution. 2004; 58: 1064-1072
-
Purcell J. Chapuisat M. Bidirectional shifts in colony queen number in a socially polymorphic ant population. Evolution. 2013; 67: 1169-1180
-
Schwander T. Rosset H. Chapuisat M. Division of labour and worker size polymorphism in ant colonies: the impact of social and genetic factors. Behav. Ecol. Sociobiol. 2005; 59: 215-221
-
Rosset H. Chapuisat M. Alternative life-histories in a socially polymorphic ant. Evol. Ecol. 2007; 21: 577-588
-
Rosset H. Chapuisat M. Sex allocation conflict in ants: when the queen rules. Curr. Biol. 2006; 16: 328-331
-
Purcell J. Chapuisat M. The influence of social structure on brood survival and development in a socially polymorphic ant: insights from a cross-fostering experiment. J. Evol. Biol. 2012; 25: 2288-2297
-
Parchman T.L. Gompert Z. Mudge J. Schilkey F.D. Benkman C.W. Buerkle C.A. Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol. Ecol. 2012; 21: 2991-3005
-
Elshire R.J. Glaubitz J.C. Sun Q. Poland J.A. Kawamoto K. Buckler E.S. Mitchell S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011; 6: e19379
-
Hauschteck-Jungen E. Jungen H. Ant chromosomes. I. The genus Formica. Insectes Soc. 1976; 23: 513-524
-
Meunier J. Chapuisat M. The determinants of queen size in a socially polymorphic ant. J. Evol. Biol. 2009; 22: 1906-1913
-
Huynh L.Y. Maney D.L. Thomas J.W. Chromosome-wide linkage disequilibrium caused by an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis). Heredity (Edinb). 2011; 106: 537-546
-
Lyon M.F. Transmission ratio distortion in mice. Annu. Rev. Genet. 2003; 37: 393-408
-
Hurst L.D. Atlan A. Bengtsson B.O. Genetic conflicts. Q. Rev. Biol. 1996; 71: 317-364
-
Keller L. Ross K.G. Selfish genes: a green beard in the red fire ant. Nature. 1998; 394: 573-575
-
Ross K.G. Keller L. Genetic control of social organization in an ant. Proc. Natl. Acad. Sci. USA. 1998; 95: 14232-14237
-
Moreau C.S. Bell C.D. Vila R. Archibald S.B. Pierce N.E. Phylogeny of the ants: diversification in the age of angiosperms. Science. 2006; 312: 101-104
-
Noor M.A.F. Grams K.L. Bertucci L.A. Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA. 2001; 98: 12084-12088
-
Hoffmann A.A. Rieseberg L.H. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation?. Annu. Rev. Ecol. Evol. Syst. 2008; 39: 21-42
-
Fishman L. Stathos A. Beardsley P.M. Williams C.F. Hill J.P. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkey flowers). Evolution. 2013; 67: 2547-2560
-
Dobzhansky T. Genetics and the Origin of Species. Third Edition. Columbia University Press, New York1951
-
Feder J.L. Nosil P. Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions?. Evolution. 2009; 63: 3061-3075
-
Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013; 14: 113-124
-
Bergero R. Charlesworth D. Filatov D.A. Moore R.C. Defining regions and rearrangements of the Silene latifolia Y chromosome. Genetics. 2008; 178: 2045-2053
-
Natri H.M. Shikano T. Merilä J. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol. Biol. Evol. 2013; 30: 1131-1144
-
Thomas J.W. Cáceres M. Lowman J.J. Morehouse C.B. Short M.E. Baldwin E.L. Maney D.L. Martin C.L. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 2008; 179: 1455-1468
-
Bennett D. The T-locus of the mouse. Cell. 1975; 6: 441-454
-
Hammer M.F. Schimenti J. Silver L.M. Evolution of mouse chromosome 17 and the origin of inversions associated with t haplotypes. Proc. Natl. Acad. Sci. USA. 1989; 86: 3261-3265
-
Feder J.L. Gejji R. Yeaman S. Nosil P. Establishment of new mutations under divergence and genome hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012; 367: 461-474
-
Renaut S. Grassa C.J. Yeaman S. Moyers B.T. Lai Z. Kane N.C. Bowers J.E. Burke J.M. Rieseberg L.H. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat. Commun. 2013; 4: 1827
-
Lowry D.B. Willis J.H. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010; 8: e1000500
Article Info
Publication History
Published: October 23, 2014 Accepted: September 24, 2014 Received in revised form: August 24, 2014 Received: April 18, 2014
Identification
DOI: https://doi.org/10.1016/j.cub.2014.09.071
Copyright
© 2014 Elsevier Ltd. Published by Elsevier Inc.
User License
ScienceDirect
Access this article on ScienceDirect